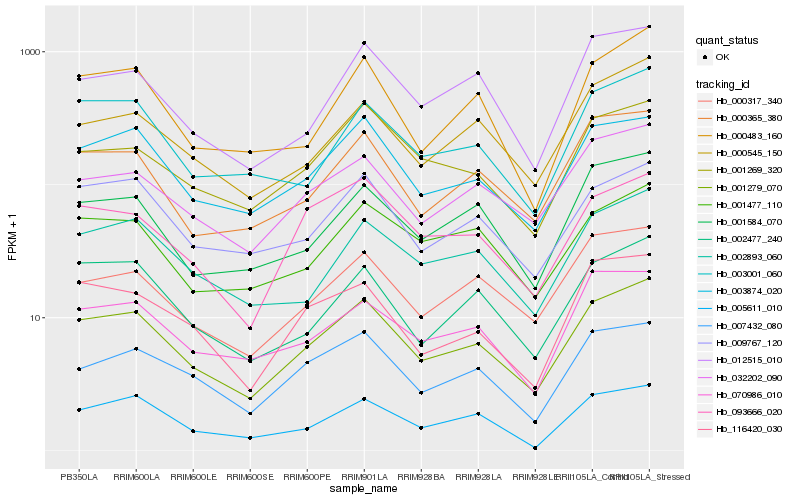
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001279_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633300 [Jatropha curcas] |
| 2 |
Hb_003874_020 |
0.072966008 |
- |
- |
hypothetical protein POPTR_0012s02280g [Populus trichocarpa] |
| 3 |
Hb_032202_090 |
0.0816204953 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Jatropha curcas] |
| 4 |
Hb_009767_120 |
0.0838632262 |
- |
- |
PREDICTED: probable prefoldin subunit 2 [Jatropha curcas] |
| 5 |
Hb_000365_380 |
0.0854611136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002893_060 |
0.0860896246 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 50 [Jatropha curcas] |
| 7 |
Hb_007432_080 |
0.0866796843 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1-like [Jatropha curcas] |
| 8 |
Hb_002477_240 |
0.0877613742 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 9 |
Hb_001584_070 |
0.0887912874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000317_340 |
0.089107325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000545_150 |
0.090543127 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |
| 12 |
Hb_012515_010 |
0.0914591816 |
- |
- |
PREDICTED: 60S ribosomal protein L30-like [Gossypium raimondii] |
| 13 |
Hb_005611_010 |
0.0922475834 |
- |
- |
PREDICTED: protein prune homolog [Jatropha curcas] |
| 14 |
Hb_001269_320 |
0.0928827852 |
- |
- |
40S ribosomal protein S12A [Hevea brasiliensis] |
| 15 |
Hb_000483_160 |
0.0929905361 |
- |
- |
PREDICTED: 60S ribosomal protein L34 [Jatropha curcas] |
| 16 |
Hb_003001_060 |
0.0940954239 |
- |
- |
wound-responsive family protein [Populus trichocarpa] |
| 17 |
Hb_093666_020 |
0.0942175288 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 18 |
Hb_070986_010 |
0.0944429192 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 19 |
Hb_116420_030 |
0.0944696658 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 20 |
Hb_001477_110 |
0.094546159 |
- |
- |
prohibitin, putative [Ricinus communis] |