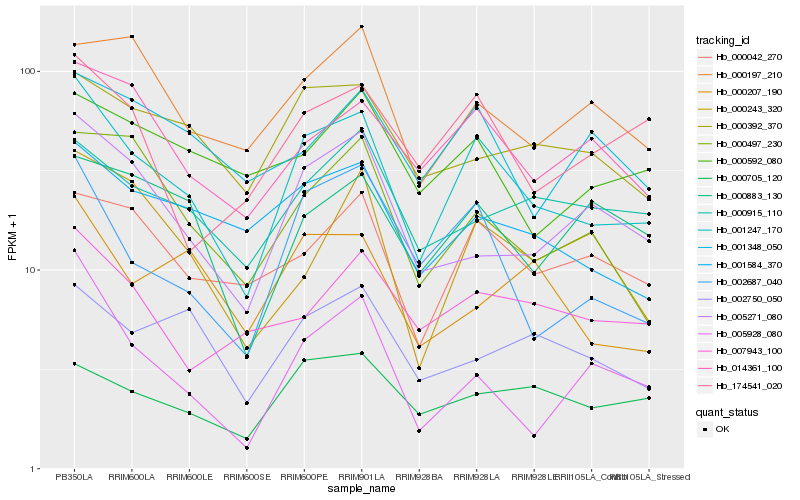
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_170 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 2 |
Hb_002687_040 |
0.1354218699 |
- |
- |
PREDICTED: alpha-galactosidase 1-like [Jatropha curcas] |
| 3 |
Hb_001584_370 |
0.147771523 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 4 |
Hb_014361_100 |
0.1480351019 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Cicer arietinum] |
| 5 |
Hb_005271_080 |
0.1515756679 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 6 |
Hb_000042_270 |
0.1520726022 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 7 |
Hb_000497_230 |
0.1588030211 |
- |
- |
PREDICTED: probable protein S-acyltransferase 6 [Jatropha curcas] |
| 8 |
Hb_000392_370 |
0.1602341125 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 9 |
Hb_174541_020 |
0.1631218857 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 6-like [Populus euphratica] |
| 10 |
Hb_000915_110 |
0.1641298057 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 11 |
Hb_000243_320 |
0.1655513235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000705_120 |
0.1666845977 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000883_130 |
0.166913889 |
- |
- |
PREDICTED: novel plant SNARE 11 [Jatropha curcas] |
| 14 |
Hb_007943_100 |
0.1687801976 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_000197_210 |
0.1692977103 |
- |
- |
phosphatidylinositol transporter, putative [Ricinus communis] |
| 16 |
Hb_002750_050 |
0.1695203346 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 17 |
Hb_001348_050 |
0.1716440146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000207_190 |
0.171884228 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 19 |
Hb_000592_080 |
0.1739724554 |
- |
- |
PREDICTED: thymidylate kinase [Jatropha curcas] |
| 20 |
Hb_005928_080 |
0.1743059047 |
- |
- |
PREDICTED: probable galacturonosyltransferase 6 isoform X1 [Jatropha curcas] |