| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
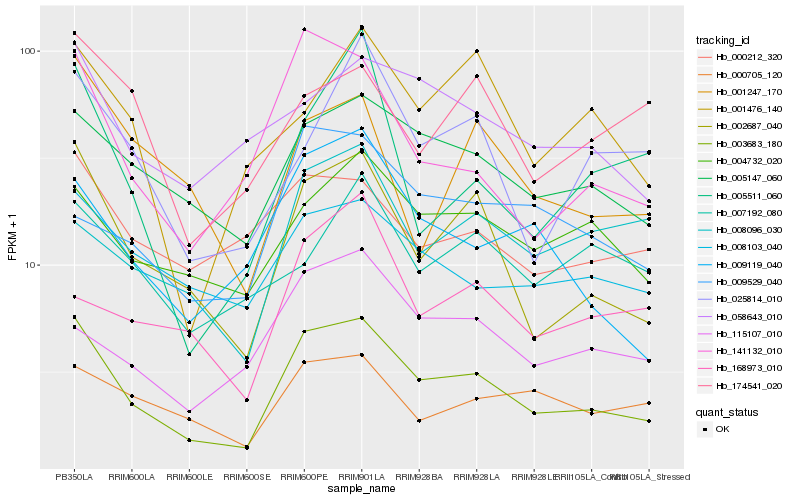
Hb_003683_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002687_040 |
0.1063427616 |
- |
- |
PREDICTED: alpha-galactosidase 1-like [Jatropha curcas] |
| 3 |
Hb_005147_060 |
0.1356653009 |
- |
- |
PREDICTED: shaggy-related protein kinase kappa [Populus euphratica] |
| 4 |
Hb_141132_010 |
0.1431017111 |
- |
- |
Hsc70-interacting protein, putative [Ricinus communis] |
| 5 |
Hb_008103_040 |
0.1516938812 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_004732_020 |
0.153034978 |
- |
- |
PREDICTED: chitobiosyldiphosphodolichol beta-mannosyltransferase isoform X1 [Populus euphratica] |
| 7 |
Hb_009119_040 |
0.1560744566 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2A [Jatropha curcas] |
| 8 |
Hb_058643_010 |
0.1627529204 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 9 |
Hb_115107_010 |
0.1653910053 |
- |
- |
Early-responsive to dehydration stress protein (ERD4) isoform 1 [Theobroma cacao] |
| 10 |
Hb_000212_320 |
0.1656807264 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 11 |
Hb_000705_120 |
0.1697342539 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_025814_010 |
0.1704303301 |
- |
- |
hypothetical protein PRUPE_ppa015585mg [Prunus persica] |
| 13 |
Hb_001247_170 |
0.1749297908 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 14 |
Hb_009529_040 |
0.1783652649 |
- |
- |
PREDICTED: transmembrane protein 120 homolog isoform X1 [Malus domestica] |
| 15 |
Hb_001476_140 |
0.1808734667 |
- |
- |
PREDICTED: uncharacterized protein LOC105630497 [Jatropha curcas] |
| 16 |
Hb_008096_030 |
0.1816970495 |
- |
- |
PREDICTED: uncharacterized protein LOC105638438 [Jatropha curcas] |
| 17 |
Hb_174541_020 |
0.1847256207 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 6-like [Populus euphratica] |
| 18 |
Hb_168973_010 |
0.1858327286 |
- |
- |
- |
| 19 |
Hb_005511_060 |
0.186245863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007192_080 |
0.186331255 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK3 isoform X2 [Jatropha curcas] |