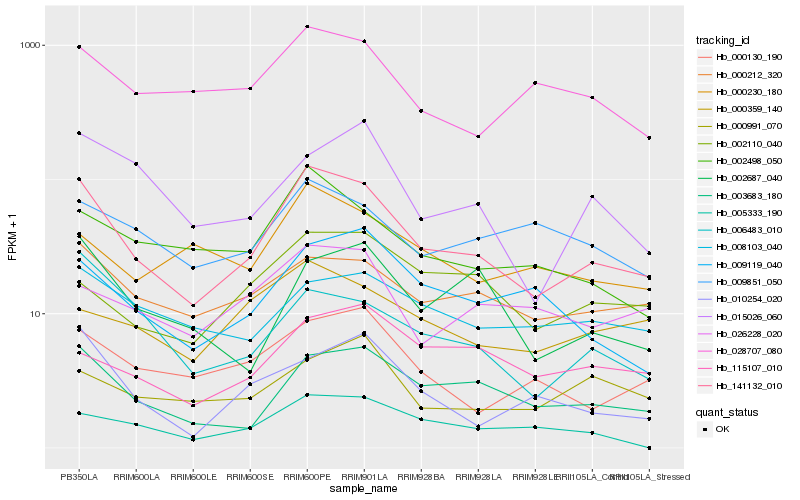
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_141132_010 |
0.0 |
- |
- |
Hsc70-interacting protein, putative [Ricinus communis] |
| 2 |
Hb_003683_180 |
0.1431017111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002498_050 |
0.1653172682 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 4 |
Hb_000212_320 |
0.1661051453 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 5 |
Hb_009119_040 |
0.1764576809 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2A [Jatropha curcas] |
| 6 |
Hb_028707_080 |
0.1774119488 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 7 |
Hb_000130_190 |
0.1788539971 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 8 |
Hb_006483_010 |
0.1848513252 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_002687_040 |
0.1850794326 |
- |
- |
PREDICTED: alpha-galactosidase 1-like [Jatropha curcas] |
| 10 |
Hb_015026_060 |
0.185527534 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 11 |
Hb_002110_040 |
0.1857350158 |
- |
- |
hypothetical protein AMTR_s00135p00074190 [Amborella trichopoda] |
| 12 |
Hb_009851_050 |
0.1866014215 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 13 |
Hb_026228_020 |
0.1868339593 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 14 |
Hb_008103_040 |
0.1871852307 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_000230_180 |
0.1883134208 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 16 |
Hb_000359_140 |
0.1895931183 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 17 |
Hb_010254_020 |
0.1898345139 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 18 |
Hb_115107_010 |
0.195081525 |
- |
- |
Early-responsive to dehydration stress protein (ERD4) isoform 1 [Theobroma cacao] |
| 19 |
Hb_005333_190 |
0.1968049035 |
- |
- |
PREDICTED: AP-4 complex subunit mu isoform X2 [Populus euphratica] |
| 20 |
Hb_000991_070 |
0.2003355208 |
- |
- |
phytochrome A specific signal transduction component family protein [Populus trichocarpa] |