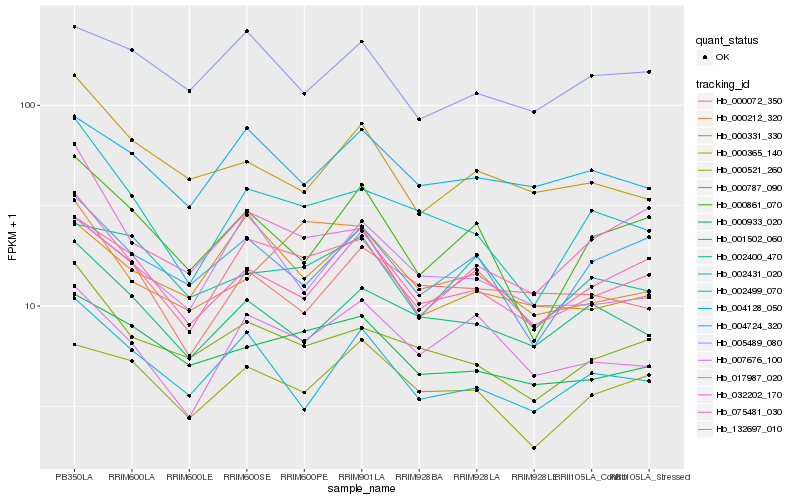
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132697_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_017987_020 |
0.0905538101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000787_090 |
0.0924915114 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Vitis vinifera] |
| 4 |
Hb_002431_020 |
0.0951411532 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 5 |
Hb_075481_030 |
0.097854317 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000933_020 |
0.0999288476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000861_070 |
0.1017612345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004724_320 |
0.1020125446 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 9 |
Hb_004128_050 |
0.106764304 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000331_330 |
0.1077707225 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X1 [Jatropha curcas] |
| 11 |
Hb_032202_170 |
0.1086964271 |
- |
- |
PREDICTED: uncharacterized protein LOC105643056 [Jatropha curcas] |
| 12 |
Hb_000212_320 |
0.1096794434 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 13 |
Hb_001502_060 |
0.1104014204 |
- |
- |
DNA mismatch repair protein MSH2, putative [Ricinus communis] |
| 14 |
Hb_002499_070 |
0.1105935337 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000521_260 |
0.1126344086 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_002400_470 |
0.1151916618 |
- |
- |
PREDICTED: calcium uptake protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_000365_140 |
0.1152863731 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 18 |
Hb_000072_350 |
0.1160524735 |
- |
- |
PREDICTED: WPP domain-interacting tail-anchored protein 1 [Jatropha curcas] |
| 19 |
Hb_005489_080 |
0.1160954046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007676_100 |
0.1172436069 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |