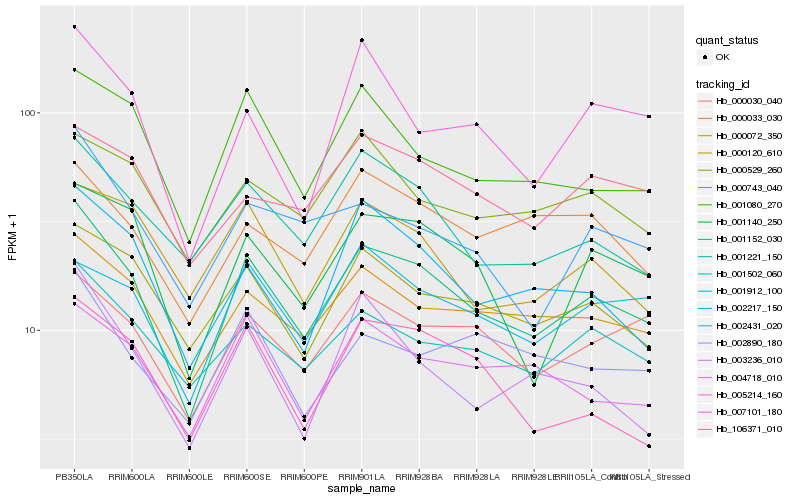
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001152_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s22050g [Populus trichocarpa] |
| 2 |
Hb_000072_350 |
0.0924428628 |
- |
- |
PREDICTED: WPP domain-interacting tail-anchored protein 1 [Jatropha curcas] |
| 3 |
Hb_001502_060 |
0.0996356465 |
- |
- |
DNA mismatch repair protein MSH2, putative [Ricinus communis] |
| 4 |
Hb_000529_260 |
0.1006775211 |
- |
- |
PREDICTED: protein SGT1 homolog At5g65490 [Jatropha curcas] |
| 5 |
Hb_001080_270 |
0.1024594328 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 6 |
Hb_001140_250 |
0.1026501622 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_001221_150 |
0.1037096936 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 8 |
Hb_002217_150 |
0.106340795 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 9 |
Hb_000030_040 |
0.1067958358 |
- |
- |
yth domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_004718_010 |
0.1095586687 |
- |
- |
PREDICTED: uncharacterized protein LOC105638290 [Jatropha curcas] |
| 11 |
Hb_002431_020 |
0.1122954366 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 12 |
Hb_001912_100 |
0.113498462 |
- |
- |
PREDICTED: uncharacterized protein LOC105636609 [Jatropha curcas] |
| 13 |
Hb_005214_160 |
0.1168704964 |
- |
- |
PREDICTED: heat shock 70 kDa protein, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000120_610 |
0.117165827 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 15 |
Hb_000033_030 |
0.117413454 |
- |
- |
PREDICTED: ankyrin repeat protein SKIP35 [Jatropha curcas] |
| 16 |
Hb_002890_180 |
0.1176145979 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3 [Jatropha curcas] |
| 17 |
Hb_003236_010 |
0.1177590374 |
- |
- |
PREDICTED: uncharacterized protein LOC105635957 [Jatropha curcas] |
| 18 |
Hb_007101_180 |
0.1177668831 |
- |
- |
PREDICTED: DNA polymerase beta [Jatropha curcas] |
| 19 |
Hb_106371_010 |
0.1183778808 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_000743_040 |
0.1185318691 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |