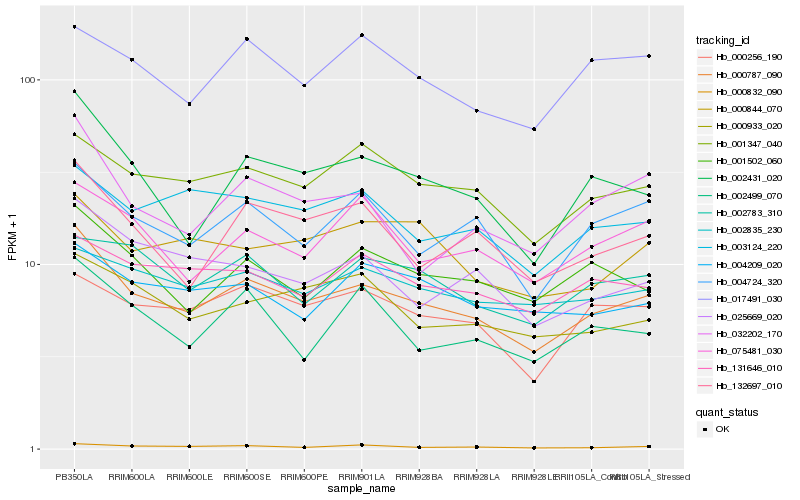
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000787_090 |
0.0 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Vitis vinifera] |
| 2 |
Hb_002431_020 |
0.0836301611 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 3 |
Hb_132697_010 |
0.0924915114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001347_040 |
0.0940675859 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 5 |
Hb_004724_320 |
0.0966721365 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 6 |
Hb_001502_060 |
0.0969551246 |
- |
- |
DNA mismatch repair protein MSH2, putative [Ricinus communis] |
| 7 |
Hb_000832_090 |
0.1001636999 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 8 |
Hb_025669_020 |
0.101483703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_032202_170 |
0.1052990539 |
- |
- |
PREDICTED: uncharacterized protein LOC105643056 [Jatropha curcas] |
| 10 |
Hb_000933_020 |
0.1056367833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000256_190 |
0.1063802973 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 12 |
Hb_003124_220 |
0.107423576 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 13 |
Hb_075481_030 |
0.1075349726 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X3 [Jatropha curcas] |
| 14 |
Hb_131646_010 |
0.1078243583 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_002499_070 |
0.1090456551 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000844_070 |
0.109480716 |
- |
- |
hypothetical protein POPTR_0014s03640g [Populus trichocarpa] |
| 17 |
Hb_002835_230 |
0.1100070909 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_017491_030 |
0.1104523931 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 19 |
Hb_004209_020 |
0.1110753628 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002783_310 |
0.111546801 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |