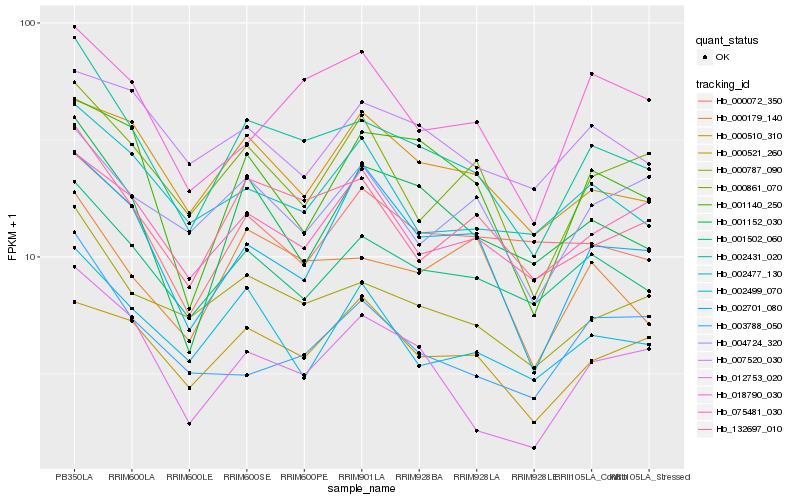
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002431_020 |
0.0 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 2 |
Hb_000787_090 |
0.0836301611 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Vitis vinifera] |
| 3 |
Hb_001502_060 |
0.0936150092 |
- |
- |
DNA mismatch repair protein MSH2, putative [Ricinus communis] |
| 4 |
Hb_132697_010 |
0.0951411532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001152_030 |
0.1122954366 |
- |
- |
hypothetical protein POPTR_0002s22050g [Populus trichocarpa] |
| 6 |
Hb_000179_140 |
0.1123802496 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_002477_130 |
0.121436697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_012753_020 |
0.1222167087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000861_070 |
0.1230951066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002499_070 |
0.1239567802 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000510_310 |
0.124336835 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 12 |
Hb_018790_030 |
0.1261337959 |
- |
- |
PREDICTED: cyclic phosphodiesterase-like [Jatropha curcas] |
| 13 |
Hb_000072_350 |
0.1267302882 |
- |
- |
PREDICTED: WPP domain-interacting tail-anchored protein 1 [Jatropha curcas] |
| 14 |
Hb_001140_250 |
0.1273793321 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_075481_030 |
0.1276067115 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X3 [Jatropha curcas] |
| 16 |
Hb_002701_080 |
0.1286233371 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like [Jatropha curcas] |
| 17 |
Hb_004724_320 |
0.1300405676 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 18 |
Hb_003788_050 |
0.1311211509 |
- |
- |
PREDICTED: interferon-induced guanylate-binding protein 2-like [Jatropha curcas] |
| 19 |
Hb_000521_260 |
0.1321662256 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_007520_030 |
0.1329045621 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |