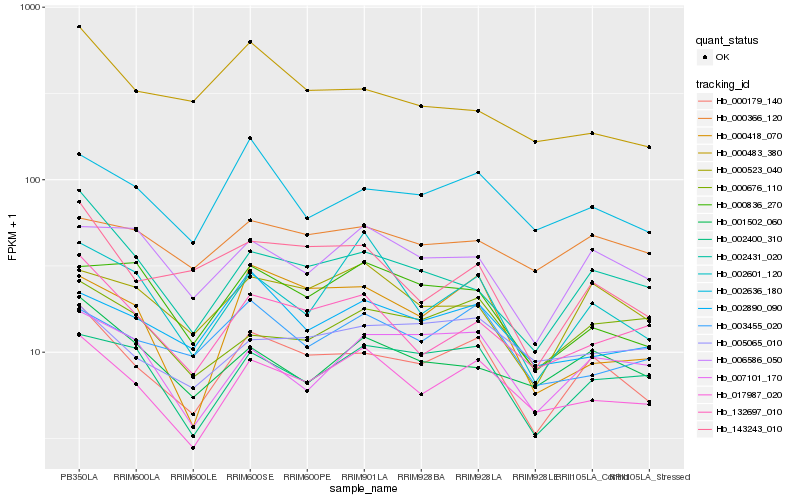
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000179_140 |
0.0 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_017987_020 |
0.1058221022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002431_020 |
0.1123802496 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 4 |
Hb_002636_180 |
0.1143404033 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 5 |
Hb_143243_010 |
0.117644757 |
- |
- |
PREDICTED: uncharacterized protein LOC105644190 [Jatropha curcas] |
| 6 |
Hb_132697_010 |
0.1196061275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002400_310 |
0.1197726898 |
- |
- |
hypothetical protein JCGZ_07138 [Jatropha curcas] |
| 8 |
Hb_000523_040 |
0.1240198537 |
- |
- |
PREDICTED: auxin response factor 8 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007101_170 |
0.1272144372 |
- |
- |
hypothetical protein POPTR_0010s10490g [Populus trichocarpa] |
| 10 |
Hb_001502_060 |
0.1285822282 |
- |
- |
DNA mismatch repair protein MSH2, putative [Ricinus communis] |
| 11 |
Hb_006586_050 |
0.1304118991 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 12 |
Hb_002890_090 |
0.1321162458 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 13 |
Hb_003455_020 |
0.1327862139 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 14 |
Hb_000676_110 |
0.1352082559 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 15 |
Hb_005065_010 |
0.1357265812 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI11 [Jatropha curcas] |
| 16 |
Hb_000418_070 |
0.1375265083 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 17 |
Hb_000836_270 |
0.1383014302 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 18 |
Hb_000366_120 |
0.138540625 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 19 |
Hb_000483_380 |
0.1407946956 |
- |
- |
PREDICTED: bax inhibitor 1-like [Jatropha curcas] |
| 20 |
Hb_002601_120 |
0.1418470961 |
- |
- |
PREDICTED: uncharacterized protein LOC105634921 [Jatropha curcas] |