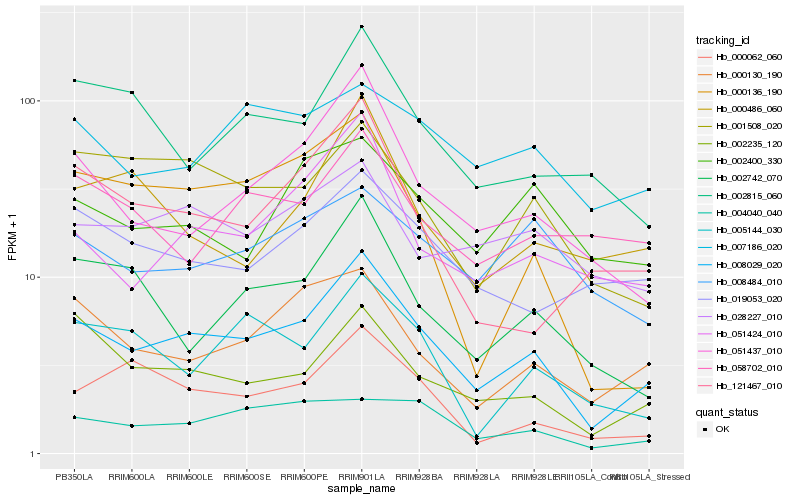
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008029_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 2 |
Hb_051437_010 |
0.1466854823 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002235_120 |
0.1502445674 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 4 |
Hb_000130_190 |
0.1507474183 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 5 |
Hb_051424_010 |
0.1540060327 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 6 |
Hb_019053_020 |
0.1604297711 |
- |
- |
hypothetical protein MIMGU_mgv1a0105441mg, partial [Erythranthe guttata] |
| 7 |
Hb_001508_020 |
0.1604524453 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 8 |
Hb_002742_070 |
0.1616392348 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 9 |
Hb_000062_060 |
0.1654875902 |
transcription factor |
TF Family: Orphans |
multi-sensor hybrid histidine kinase [Plautia stali symbiont] |
| 10 |
Hb_002815_060 |
0.1657422868 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 11 |
Hb_121467_010 |
0.1701387716 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 12 |
Hb_028227_010 |
0.1702102401 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 13 |
Hb_004040_040 |
0.1706186253 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 14 |
Hb_005144_030 |
0.1709498303 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 15 |
Hb_000486_060 |
0.1738137915 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like isoform X2 [Glycine max] |
| 16 |
Hb_007186_020 |
0.1739518656 |
- |
- |
Eukaryotic initiation factor 4A-3 [Gossypium arboreum] |
| 17 |
Hb_000136_190 |
0.1743943365 |
- |
- |
PREDICTED: protein-S-isoprenylcysteine O-methyltransferase A-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_058702_010 |
0.1749470549 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_008484_010 |
0.1756024967 |
- |
- |
hypothetical protein B456_008G050700 [Gossypium raimondii] |
| 20 |
Hb_002400_330 |
0.1757703346 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |