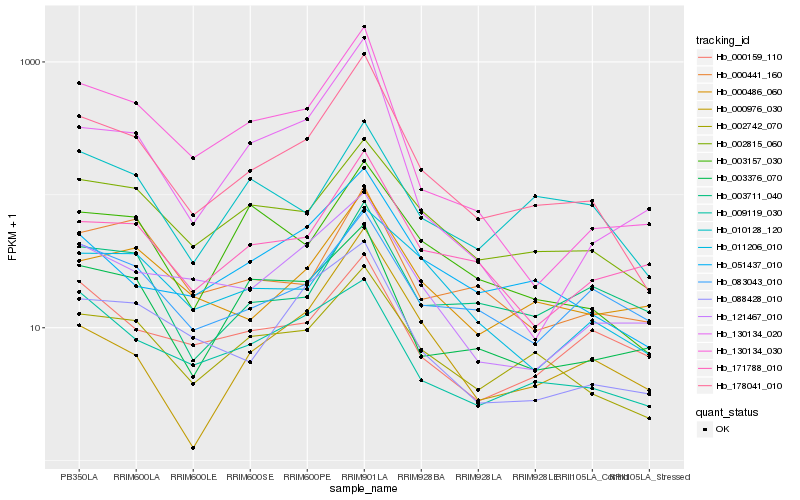
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_178041_010 |
0.0 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Populus euphratica] |
| 2 |
Hb_002742_070 |
0.1199584266 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 3 |
Hb_051437_010 |
0.1385336454 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 4 |
Hb_130134_030 |
0.1457929747 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_14472 [Jatropha curcas] |
| 5 |
Hb_002815_060 |
0.1466903329 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 6 |
Hb_088428_010 |
0.1505171347 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 7 |
Hb_171788_010 |
0.1564335438 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 8 |
Hb_000486_060 |
0.1598793493 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like isoform X2 [Glycine max] |
| 9 |
Hb_121467_010 |
0.1606400467 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 10 |
Hb_003157_030 |
0.1647583299 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Nicotiana sylvestris] |
| 11 |
Hb_003376_070 |
0.1679598443 |
- |
- |
hypothetical protein B456_009G419800 [Gossypium raimondii] |
| 12 |
Hb_083043_010 |
0.171200002 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 13 |
Hb_011206_010 |
0.1722294729 |
- |
- |
PREDICTED: something about silencing protein 10 isoform X2 [Vitis vinifera] |
| 14 |
Hb_009119_030 |
0.1726739593 |
- |
- |
GMFP4 [Medicago truncatula] |
| 15 |
Hb_000441_160 |
0.1757942769 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 16 |
Hb_000976_030 |
0.1771100227 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like [Citrus sinensis] |
| 17 |
Hb_003711_040 |
0.1790502367 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 18 |
Hb_130134_020 |
0.1791209592 |
- |
- |
- |
| 19 |
Hb_000159_110 |
0.1856222471 |
- |
- |
PREDICTED: thioredoxin-like protein YLS8 isoform X2 [Malus domestica] |
| 20 |
Hb_010128_120 |
0.1858150637 |
- |
- |
hypothetical protein JCGZ_23167 [Jatropha curcas] |