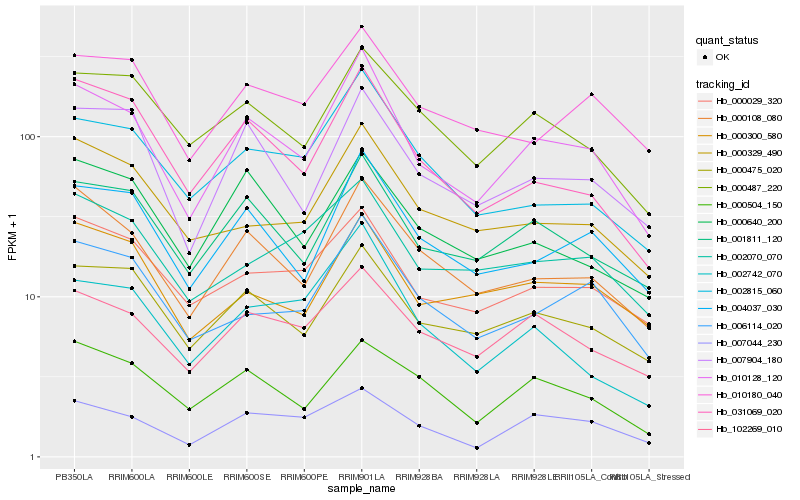
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010128_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_23167 [Jatropha curcas] |
| 2 |
Hb_000108_080 |
0.0948128249 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 3 |
Hb_001811_120 |
0.0954779254 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004037_030 |
0.095628858 |
- |
- |
PREDICTED: DNA repair endonuclease UVH1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000475_020 |
0.107052626 |
- |
- |
PREDICTED: dehydrogenase/reductase SDR family member on chromosome X homolog isoform X2 [Jatropha curcas] |
| 6 |
Hb_007904_180 |
0.1105011609 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Jatropha curcas] |
| 7 |
Hb_031069_020 |
0.1116639081 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 3-like [Jatropha curcas] |
| 8 |
Hb_102269_010 |
0.1124170537 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 9 |
Hb_006114_020 |
0.1131982618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007044_230 |
0.1157428531 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 11 |
Hb_000029_320 |
0.1174324059 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 12 |
Hb_000487_220 |
0.1185588288 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 13 |
Hb_002815_060 |
0.1192608444 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 14 |
Hb_000504_150 |
0.1239195513 |
- |
- |
PREDICTED: nuclear pore complex protein GP210 [Jatropha curcas] |
| 15 |
Hb_010180_040 |
0.1241327392 |
- |
- |
PREDICTED: prosaposin isoform X1 [Jatropha curcas] |
| 16 |
Hb_000329_490 |
0.1243041897 |
- |
- |
hypothetical protein JCGZ_14582 [Jatropha curcas] |
| 17 |
Hb_002742_070 |
0.1270905081 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 18 |
Hb_000300_580 |
0.1272161758 |
- |
- |
PREDICTED: importin-5-like [Jatropha curcas] |
| 19 |
Hb_000640_200 |
0.1290842381 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |
| 20 |
Hb_002070_070 |
0.1293872173 |
- |
- |
ATP binding protein, putative [Ricinus communis] |