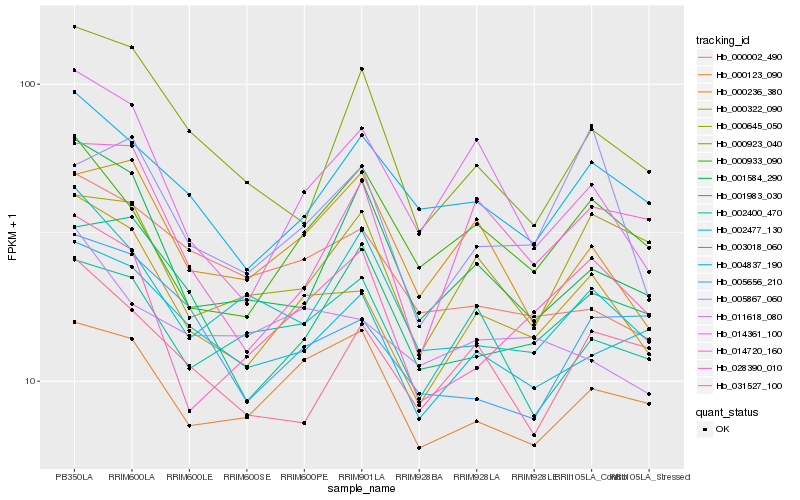
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000322_090 |
0.0 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 2 |
Hb_003018_060 |
0.0995724973 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 3 |
Hb_004837_190 |
0.1062011624 |
- |
- |
PREDICTED: uncharacterized protein LOC105648286 [Jatropha curcas] |
| 4 |
Hb_014361_100 |
0.1074519003 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Cicer arietinum] |
| 5 |
Hb_000123_090 |
0.1108934509 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 6 |
Hb_028390_010 |
0.1123220942 |
- |
- |
PREDICTED: filament-like plant protein 3 [Populus euphratica] |
| 7 |
Hb_001584_290 |
0.1128381508 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 8 |
Hb_001983_030 |
0.1155330503 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 12 isoform X2 [Jatropha curcas] |
| 9 |
Hb_031527_100 |
0.1157726673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005656_210 |
0.1157750542 |
- |
- |
PREDICTED: uncharacterized protein LOC105637440 [Jatropha curcas] |
| 11 |
Hb_014720_160 |
0.1159138061 |
- |
- |
PREDICTED: anthranilate phosphoribosyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002477_130 |
0.116778214 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000002_490 |
0.1171410753 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 14 |
Hb_011618_080 |
0.1181846048 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 15 |
Hb_000933_090 |
0.1183185412 |
- |
- |
PREDICTED: probable protein S-acyltransferase 22 [Jatropha curcas] |
| 16 |
Hb_000645_050 |
0.1185569919 |
- |
- |
peptidyl-prolyl cis-trans isomerase cyclophilin-type family protein [Populus trichocarpa] |
| 17 |
Hb_000236_380 |
0.1196966931 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 18 |
Hb_002400_470 |
0.1200400472 |
- |
- |
PREDICTED: calcium uptake protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_000923_040 |
0.1203164016 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 20 |
Hb_005867_060 |
0.122556324 |
- |
- |
PREDICTED: NADPH--cytochrome P450 reductase [Jatropha curcas] |