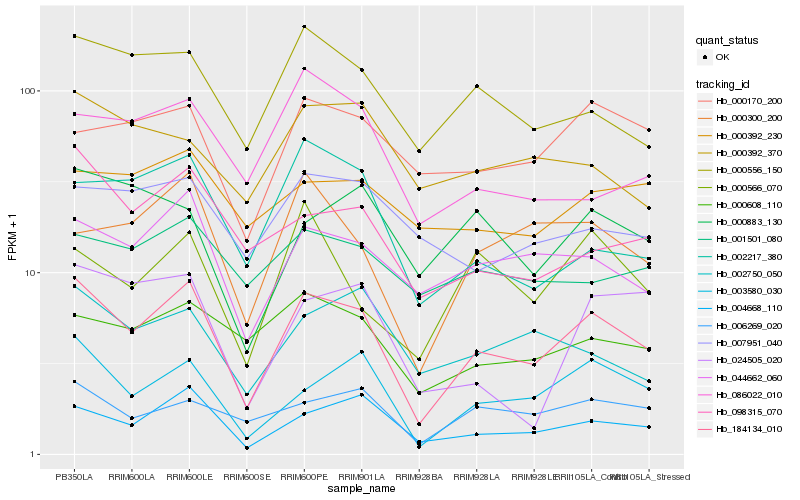
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_184134_010 |
0.0 |
- |
- |
PREDICTED: variable charge X-linked protein 3B [Jatropha curcas] |
| 2 |
Hb_004668_110 |
0.1312947502 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003580_030 |
0.1326002996 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 4 |
Hb_044662_060 |
0.1461519123 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000556_150 |
0.1575624951 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 6 |
Hb_006269_020 |
0.1580312077 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 7 |
Hb_098315_070 |
0.1619289203 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105636030 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000608_110 |
0.1662306642 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 9 |
Hb_000300_200 |
0.166591407 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_002750_050 |
0.172001089 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 11 |
Hb_002217_380 |
0.1739283967 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 12 |
Hb_000170_200 |
0.1772299892 |
- |
- |
PREDICTED: protein KRTCAP2 homolog [Jatropha curcas] |
| 13 |
Hb_000566_070 |
0.1774361432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000883_130 |
0.1775925675 |
- |
- |
PREDICTED: novel plant SNARE 11 [Jatropha curcas] |
| 15 |
Hb_001501_080 |
0.1826889484 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007951_040 |
0.1839817288 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 17 |
Hb_024505_020 |
0.1841150107 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 18 |
Hb_000392_370 |
0.1863854685 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 19 |
Hb_086022_010 |
0.1868678671 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 20 |
Hb_000392_230 |
0.1873750679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |