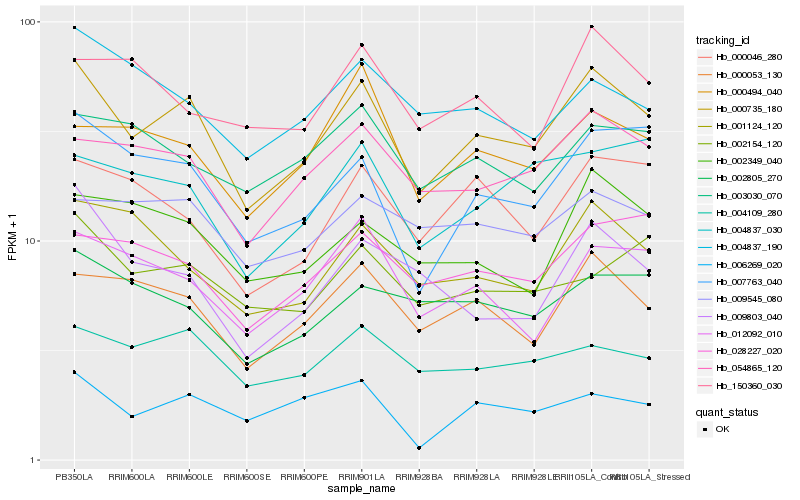
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000735_180 |
0.0 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000053_130 |
0.1008403829 |
- |
- |
PREDICTED: uncharacterized protein At1g26090, chloroplastic [Jatropha curcas] |
| 3 |
Hb_054865_120 |
0.1030137178 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 4 |
Hb_007763_040 |
0.1054664439 |
- |
- |
- |
| 5 |
Hb_004109_280 |
0.1096729978 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 6 |
Hb_012092_010 |
0.1165218137 |
- |
- |
PREDICTED: uncharacterized protein LOC105635929 [Jatropha curcas] |
| 7 |
Hb_001124_120 |
0.1184040188 |
- |
- |
PREDICTED: uncharacterized protein LOC105631831 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002349_040 |
0.1205767619 |
- |
- |
PREDICTED: RNA pseudouridine synthase 5 [Jatropha curcas] |
| 9 |
Hb_000046_280 |
0.1212331898 |
- |
- |
PREDICTED: uncharacterized protein LOC105631831 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006269_020 |
0.1220639774 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 11 |
Hb_004837_030 |
0.1234964024 |
- |
- |
PREDICTED: mitochondrial tRNA-specific 2-thiouridylase 1 [Jatropha curcas] |
| 12 |
Hb_028227_020 |
0.1235237432 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB isoform X1 [Jatropha curcas] |
| 13 |
Hb_002805_270 |
0.1241160528 |
- |
- |
PREDICTED: phytochromobilin:ferredoxin oxidoreductase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_009545_080 |
0.1248565178 |
- |
- |
hypothetical protein POPTR_0008s15400g [Populus trichocarpa] |
| 15 |
Hb_004837_190 |
0.1250811859 |
- |
- |
PREDICTED: uncharacterized protein LOC105648286 [Jatropha curcas] |
| 16 |
Hb_002154_120 |
0.1250997013 |
- |
- |
PREDICTED: uncharacterized protein LOC105630563 [Jatropha curcas] |
| 17 |
Hb_003030_070 |
0.1256586459 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 18 |
Hb_000494_040 |
0.1271672626 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 19 |
Hb_009803_040 |
0.1284960371 |
- |
- |
tetratricopeptide repeat-containing family protein [Populus trichocarpa] |
| 20 |
Hb_150360_030 |
0.1287499726 |
- |
- |
PREDICTED: F-box/WD-40 repeat-containing protein At3g52030-like [Jatropha curcas] |