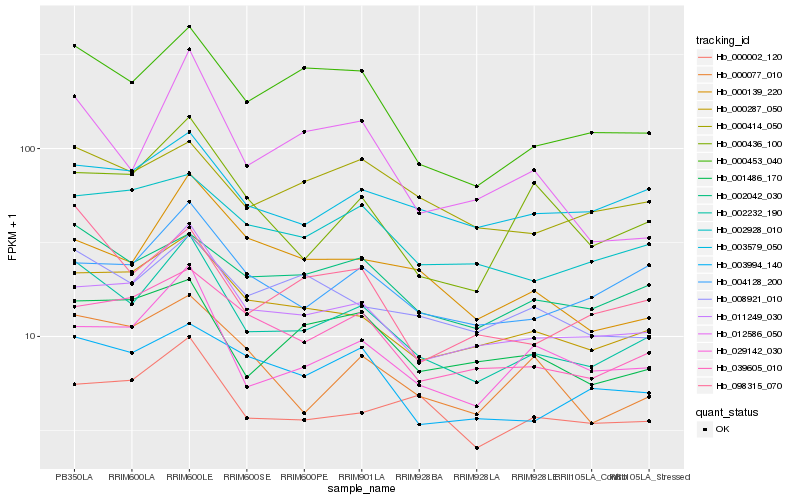
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002232_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011249_030 |
0.0936724816 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 3 |
Hb_000287_050 |
0.09586801 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 4 |
Hb_000453_040 |
0.0970333573 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 5 |
Hb_029142_030 |
0.1066348524 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 6 |
Hb_002042_030 |
0.1111200305 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 7 |
Hb_098315_070 |
0.1112612688 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105636030 isoform X1 [Jatropha curcas] |
| 8 |
Hb_039605_010 |
0.1120391848 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008921_010 |
0.1163809945 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 10 |
Hb_004128_200 |
0.1180302939 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 11 |
Hb_000077_010 |
0.1192359752 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_002928_010 |
0.1195696784 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 13 |
Hb_003994_140 |
0.1204027013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001486_170 |
0.1227961424 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 15 |
Hb_000002_120 |
0.1232870984 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 16 |
Hb_003579_050 |
0.1238144985 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 17 |
Hb_000139_220 |
0.1242398687 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_012586_050 |
0.1257584857 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Prunus mume] |
| 19 |
Hb_000414_050 |
0.1281569991 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 20 |
Hb_000436_100 |
0.1297700193 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |