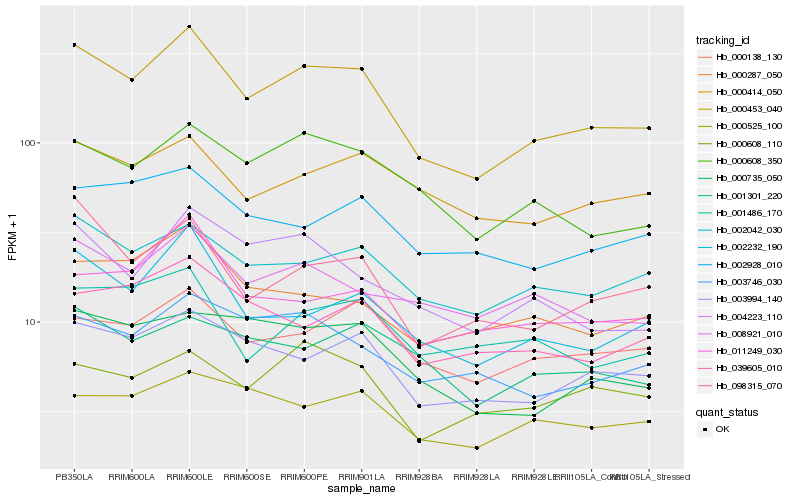
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_040 |
0.0 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 2 |
Hb_003994_140 |
0.0898554737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002232_190 |
0.0970333573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002042_030 |
0.099192007 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 5 |
Hb_000608_350 |
0.1018404106 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 6 |
Hb_098315_070 |
0.1043938913 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105636030 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000138_130 |
0.1087580624 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |
| 8 |
Hb_000735_050 |
0.1090077271 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 9 |
Hb_008921_010 |
0.1092463137 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 10 |
Hb_004223_110 |
0.109980801 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 11 |
Hb_003746_030 |
0.1103739214 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 12 |
Hb_000287_050 |
0.1117302527 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 13 |
Hb_039605_010 |
0.1143215982 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 14 |
Hb_011249_030 |
0.115001877 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 15 |
Hb_002928_010 |
0.115491264 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 16 |
Hb_000414_050 |
0.1162621182 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 17 |
Hb_000525_100 |
0.1168918314 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 18 |
Hb_001486_170 |
0.1176284319 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 19 |
Hb_001301_220 |
0.118283986 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 20 |
Hb_000608_110 |
0.1184953701 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |