| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
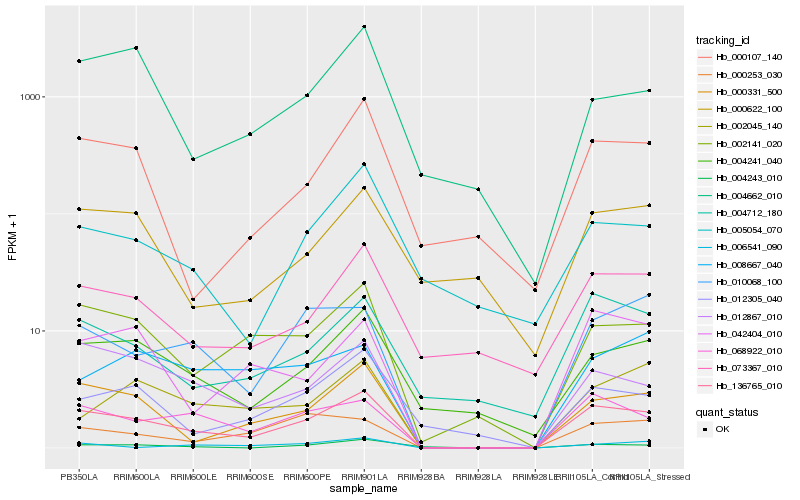
Hb_136765_010 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000331_500 |
0.1494340541 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_068922_010 |
0.1524169906 |
- |
- |
hypothetical protein POPTR_0005s17480g [Populus trichocarpa] |
| 4 |
Hb_002141_020 |
0.156156616 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 5 |
Hb_012867_010 |
0.1606883646 |
- |
- |
Vacuolar protein sorting 45 [Theobroma cacao] |
| 6 |
Hb_010068_100 |
0.1827127692 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004241_040 |
0.1846948033 |
- |
- |
- |
| 8 |
Hb_042404_010 |
0.1887762753 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 9 |
Hb_004712_180 |
0.1923088206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000253_030 |
0.2035325607 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 11 |
Hb_073367_010 |
0.2060513727 |
- |
- |
PREDICTED: origin of replication complex subunit 2 [Jatropha curcas] |
| 12 |
Hb_012305_040 |
0.2066921289 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 9A-like [Jatropha curcas] |
| 13 |
Hb_004243_010 |
0.2095448595 |
- |
- |
hypothetical protein JCGZ_01580 [Jatropha curcas] |
| 14 |
Hb_002045_140 |
0.2115091541 |
- |
- |
histone acetyltransferase, partial [Triticum aestivum] |
| 15 |
Hb_000107_140 |
0.2122631038 |
- |
- |
- |
| 16 |
Hb_008667_040 |
0.2198212352 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 17 |
Hb_006541_090 |
0.2229797603 |
- |
- |
hypothetical protein VITISV_020777 [Vitis vinifera] |
| 18 |
Hb_004662_010 |
0.2279578635 |
- |
- |
Nascent polypeptide-associated complex subunit beta [Medicago truncatula] |
| 19 |
Hb_005054_070 |
0.2309863441 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_000622_100 |
0.2309933501 |
- |
- |
PREDICTED: pre-rRNA-processing protein RIX1-like [Jatropha curcas] |