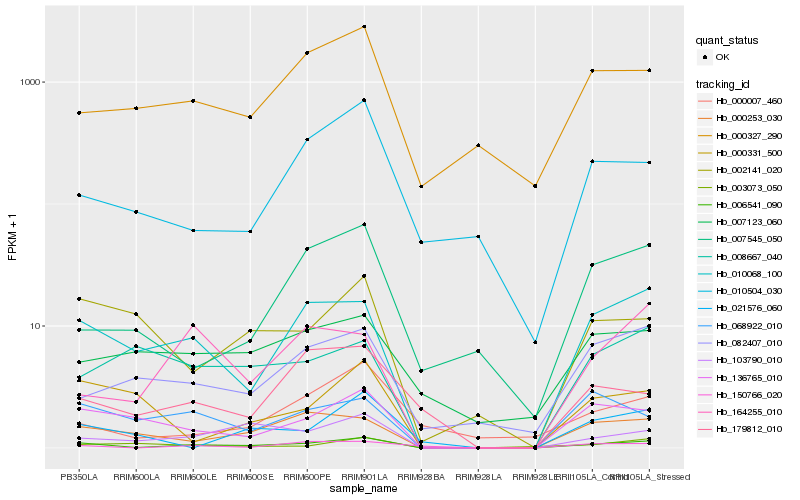
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006541_090 |
0.0 |
- |
- |
hypothetical protein VITISV_020777 [Vitis vinifera] |
| 2 |
Hb_010068_100 |
0.1990828538 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_103790_010 |
0.2225056369 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X4 [Jatropha curcas] |
| 4 |
Hb_136765_010 |
0.2229797603 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000253_030 |
0.2332324745 |
- |
- |
hypothetical protein JCGZ_23039 [Jatropha curcas] |
| 6 |
Hb_068922_010 |
0.255931503 |
- |
- |
hypothetical protein POPTR_0005s17480g [Populus trichocarpa] |
| 7 |
Hb_000327_290 |
0.2561172118 |
- |
- |
Histone H4 [Triticum urartu] |
| 8 |
Hb_002141_020 |
0.2567830093 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 9 |
Hb_150766_020 |
0.2592623881 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 10 |
Hb_082407_010 |
0.2627514514 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 11 |
Hb_164255_010 |
0.2647193957 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 12 |
Hb_010504_030 |
0.2701407133 |
- |
- |
calmodulin homolog {EST} [Brassica napus, Naehan, root, Peptide Partial, 53 aa] |
| 13 |
Hb_021576_060 |
0.2713878274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007123_060 |
0.2736695604 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 15 |
Hb_179812_010 |
0.2746377575 |
- |
- |
TdcA1-ORF1-ORF2 [Daucus carota] |
| 16 |
Hb_008667_040 |
0.2759824119 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 17 |
Hb_003073_050 |
0.2772346553 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 18 |
Hb_000007_460 |
0.2783327947 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 19 |
Hb_000331_500 |
0.2786746824 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_007545_050 |
0.2797877052 |
- |
- |
- |