| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
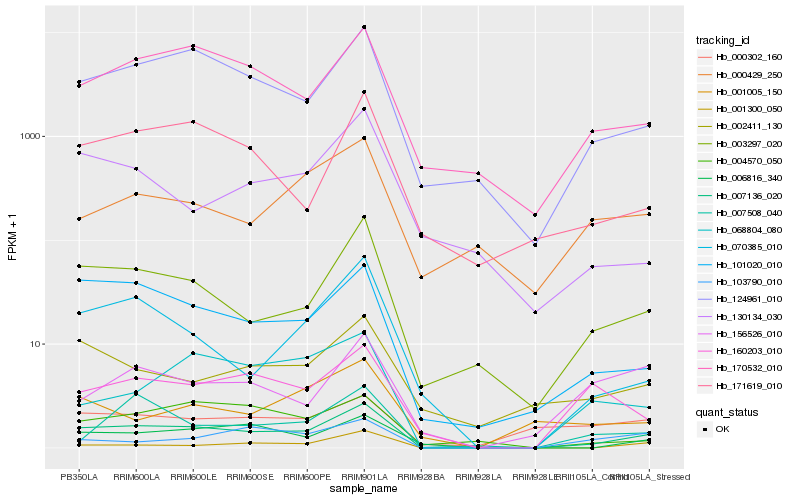
Hb_007136_020 |
0.0 |
- |
- |
PREDICTED: oxygen-dependent coproporphyrinogen-III oxidase, chloroplastic [Populus euphratica] |
| 2 |
Hb_124961_010 |
0.1623509064 |
- |
- |
hypothetical protein EUGRSUZ_B03893 [Eucalyptus grandis] |
| 3 |
Hb_003297_020 |
0.1747687786 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 4 |
Hb_101020_010 |
0.1784205997 |
- |
- |
- |
| 5 |
Hb_170532_010 |
0.1887353799 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 6 |
Hb_006816_340 |
0.1991100719 |
- |
- |
- |
| 7 |
Hb_070385_010 |
0.2089295969 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 8 |
Hb_001300_050 |
0.2100845524 |
- |
- |
- |
| 9 |
Hb_171619_010 |
0.2138282903 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_068804_080 |
0.2176023642 |
- |
- |
PREDICTED: protein BRICK 1 [Jatropha curcas] |
| 11 |
Hb_160203_010 |
0.2189905318 |
- |
- |
- |
| 12 |
Hb_004570_050 |
0.2207954503 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_002411_130 |
0.2293208921 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Populus euphratica] |
| 14 |
Hb_156526_010 |
0.2375640848 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 15 |
Hb_103790_010 |
0.2393631932 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X4 [Jatropha curcas] |
| 16 |
Hb_000302_160 |
0.2424129151 |
- |
- |
PREDICTED: uncharacterized protein LOC105648403 [Jatropha curcas] |
| 17 |
Hb_000429_250 |
0.2425113732 |
- |
- |
hypothetical protein L484_000744 [Morus notabilis] |
| 18 |
Hb_130134_030 |
0.2447498565 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_14472 [Jatropha curcas] |
| 19 |
Hb_007508_040 |
0.2460610621 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001005_150 |
0.246481465 |
- |
- |
- |