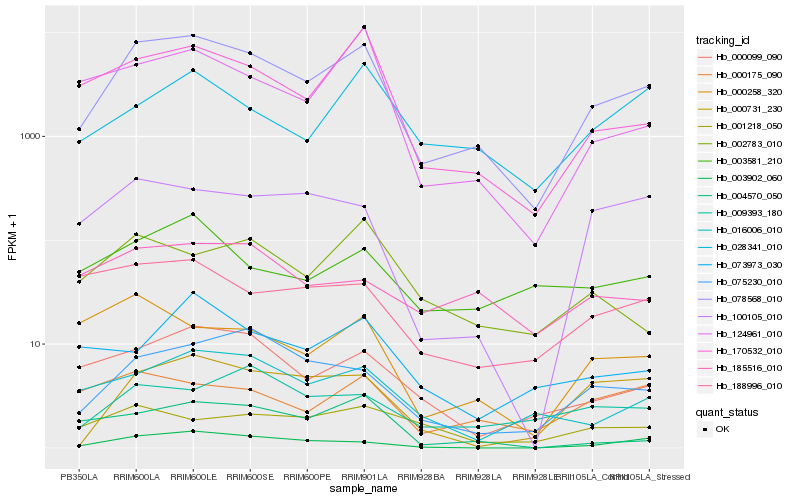
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_078568_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa013245mg [Prunus persica] |
| 2 |
Hb_170532_010 |
0.1768044106 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 3 |
Hb_000099_090 |
0.1830372007 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 4 |
Hb_075230_010 |
0.1906545936 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like [Glycine max] |
| 5 |
Hb_016006_010 |
0.191411212 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_003902_060 |
0.1944594469 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 7 |
Hb_000731_230 |
0.1995851324 |
- |
- |
PREDICTED: abnormal spindle-like microcephaly-associated protein homolog [Jatropha curcas] |
| 8 |
Hb_000175_090 |
0.2071501048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_028341_010 |
0.2087270739 |
- |
- |
hypothetical protein CICLE_v10026841mg [Citrus clementina] |
| 10 |
Hb_124961_010 |
0.2093467445 |
- |
- |
hypothetical protein EUGRSUZ_B03893 [Eucalyptus grandis] |
| 11 |
Hb_100105_010 |
0.2096828281 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 12 |
Hb_002783_010 |
0.2103720729 |
- |
- |
PREDICTED: uncharacterized protein LOC105635318 [Jatropha curcas] |
| 13 |
Hb_188996_010 |
0.2119090772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004570_050 |
0.2122774508 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_003581_210 |
0.212321867 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-binding protein WHY1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_009393_180 |
0.214892669 |
- |
- |
PREDICTED: uncharacterized protein LOC101292333 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_000258_320 |
0.2155814063 |
- |
- |
- |
| 18 |
Hb_073973_030 |
0.2217369838 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 19 |
Hb_185516_010 |
0.2219091737 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 20 |
Hb_001218_050 |
0.2239738892 |
- |
- |
Ribosome-binding protein, putative [Ricinus communis] |