| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
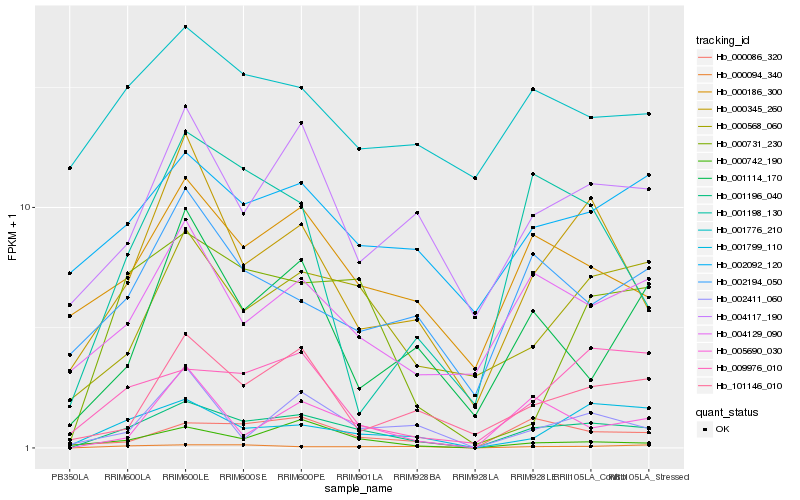
Hb_001196_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 2 |
Hb_000345_260 |
0.2024643366 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Citrus sinensis] |
| 3 |
Hb_000086_320 |
0.2025210713 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 4 |
Hb_000731_230 |
0.2061123476 |
- |
- |
PREDICTED: abnormal spindle-like microcephaly-associated protein homolog [Jatropha curcas] |
| 5 |
Hb_000094_340 |
0.207654701 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 6 |
Hb_000186_300 |
0.2085192542 |
- |
- |
PREDICTED: malonyl-CoA decarboxylase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000568_060 |
0.2131919217 |
- |
- |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 8 |
Hb_001799_110 |
0.2141910399 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_004117_190 |
0.2144513819 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 10 |
Hb_004129_090 |
0.2171894359 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 11 |
Hb_000742_190 |
0.221541856 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_101146_010 |
0.2238273439 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 13 |
Hb_009976_010 |
0.226371399 |
- |
- |
PREDICTED: uncharacterized protein LOC105630216 [Jatropha curcas] |
| 14 |
Hb_002194_050 |
0.2287137174 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002092_120 |
0.2319839818 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005690_030 |
0.2346542049 |
- |
- |
hypothetical protein VITISV_020378 [Vitis vinifera] |
| 17 |
Hb_001114_170 |
0.2381377357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002411_060 |
0.2404700704 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 19 |
Hb_001198_130 |
0.2417885591 |
- |
- |
PREDICTED: potassium channel AKT2/3 [Jatropha curcas] |
| 20 |
Hb_001776_210 |
0.2437198604 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |