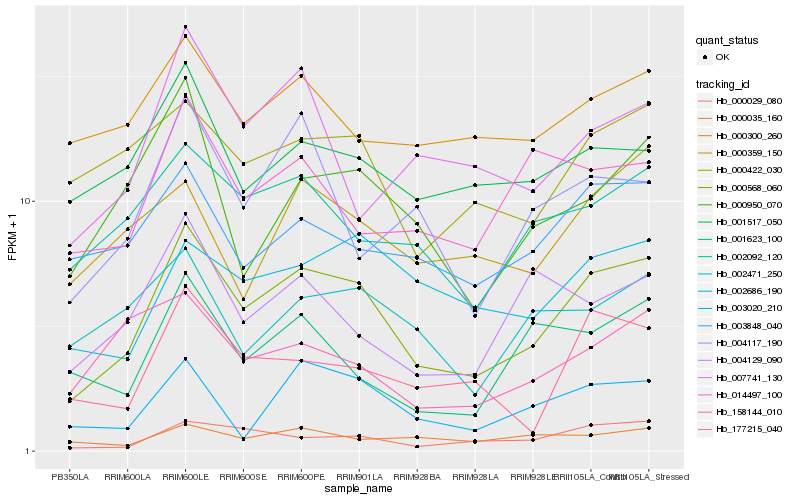
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000568_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 2 |
Hb_000029_080 |
0.1485296548 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 3 |
Hb_002092_120 |
0.1522183374 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002686_190 |
0.1602443364 |
- |
- |
- |
| 5 |
Hb_000950_070 |
0.1605963071 |
- |
- |
PREDICTED: uncharacterized protein LOC105645259 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003848_040 |
0.165078984 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 7 |
Hb_003020_210 |
0.1652081774 |
- |
- |
Structural maintenance of chromosomes 2-2 -like protein [Gossypium arboreum] |
| 8 |
Hb_000422_030 |
0.1653625081 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 9 |
Hb_177215_040 |
0.1666048628 |
- |
- |
PREDICTED: DET1- and DDB1-associated protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004117_190 |
0.1687301086 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 11 |
Hb_004129_090 |
0.169781261 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 12 |
Hb_007741_130 |
0.1708288559 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 13 |
Hb_001517_050 |
0.1710024601 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002471_250 |
0.1712011293 |
- |
- |
Tapt1/CMV receptor, putative isoform 2 [Theobroma cacao] |
| 15 |
Hb_000300_260 |
0.1729009805 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 16 |
Hb_000035_160 |
0.1732131737 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 17 |
Hb_158144_010 |
0.1735109602 |
- |
- |
- |
| 18 |
Hb_014497_100 |
0.1738798029 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000359_150 |
0.174229525 |
- |
- |
PREDICTED: protein Mpv17-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001623_100 |
0.1749103851 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |