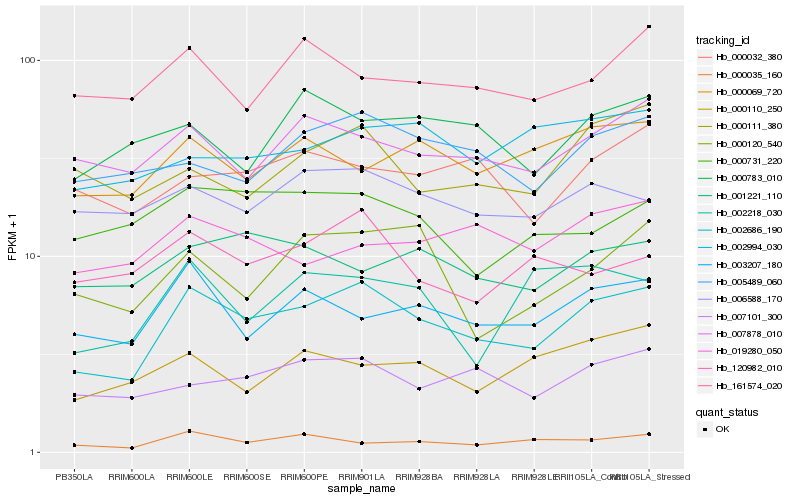
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_190 |
0.0 |
- |
- |
- |
| 2 |
Hb_003207_180 |
0.130102013 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007101_300 |
0.1307700821 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 4 |
Hb_005489_060 |
0.1329754827 |
- |
- |
Anamorsin, putative [Ricinus communis] |
| 5 |
Hb_007878_010 |
0.1332379709 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 4 [Jatropha curcas] |
| 6 |
Hb_006588_170 |
0.141943718 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |
| 7 |
Hb_000032_380 |
0.1421756537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000069_720 |
0.1422802921 |
- |
- |
PREDICTED: exosome complex component RRP41-like [Jatropha curcas] |
| 9 |
Hb_000120_540 |
0.1430895251 |
- |
- |
translation initiation factor, putative [Ricinus communis] |
| 10 |
Hb_000035_160 |
0.1446032948 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 11 |
Hb_000111_380 |
0.144948138 |
- |
- |
PREDICTED: cilia- and flagella-associated protein 20 [Jatropha curcas] |
| 12 |
Hb_001221_110 |
0.1450059278 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 13 |
Hb_161574_020 |
0.1466789423 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 14 |
Hb_120982_010 |
0.1479395938 |
- |
- |
hypothetical protein CICLE_v10003116mg, partial [Citrus clementina] |
| 15 |
Hb_002994_030 |
0.1481232794 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-like [Pyrus x bretschneideri] |
| 16 |
Hb_000783_010 |
0.1481746715 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 17 |
Hb_002218_030 |
0.1481805357 |
- |
- |
hypothetical protein B456_007G345200 [Gossypium raimondii] |
| 18 |
Hb_000110_250 |
0.1482110857 |
- |
- |
PREDICTED: homologous-pairing protein 2 homolog [Jatropha curcas] |
| 19 |
Hb_000731_220 |
0.1489776493 |
- |
- |
PREDICTED: derlin-2.2 [Jatropha curcas] |
| 20 |
Hb_019280_050 |
0.1496626709 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 2 [Jatropha curcas] |