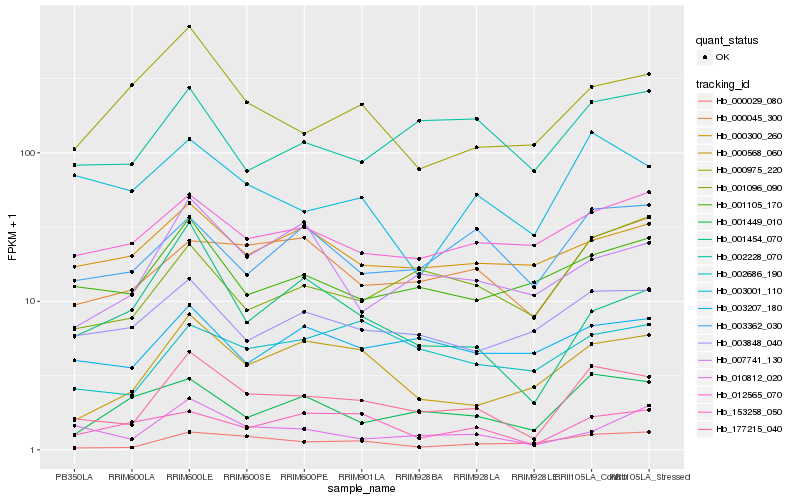
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_177215_040 |
0.0 |
- |
- |
PREDICTED: DET1- and DDB1-associated protein 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000568_060 |
0.1666048628 |
- |
- |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 3 |
Hb_000029_080 |
0.1722353912 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 4 |
Hb_000045_300 |
0.1726789191 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 5 |
Hb_001449_010 |
0.181669621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003848_040 |
0.1831076328 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 7 |
Hb_001105_170 |
0.1833397546 |
- |
- |
PREDICTED: uncharacterized protein LOC105642727 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003362_030 |
0.1834825477 |
- |
- |
hypothetical protein POPTR_0017s08440g [Populus trichocarpa] |
| 9 |
Hb_002228_070 |
0.1843409111 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 10 |
Hb_003001_110 |
0.1858685977 |
- |
- |
oleoyl-acyl carrier protein thioesterase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001096_090 |
0.1865886304 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 12 |
Hb_012565_070 |
0.1883020116 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 13 |
Hb_010812_020 |
0.1904943079 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 14 |
Hb_153258_050 |
0.1910024998 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 15 |
Hb_003207_180 |
0.1911809977 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007741_130 |
0.1917876027 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 17 |
Hb_001454_070 |
0.1929980635 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 18 |
Hb_000975_220 |
0.1932519116 |
- |
- |
Histone H2B [Medicago truncatula] |
| 19 |
Hb_002686_190 |
0.1941938411 |
- |
- |
- |
| 20 |
Hb_000300_260 |
0.1954036753 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |