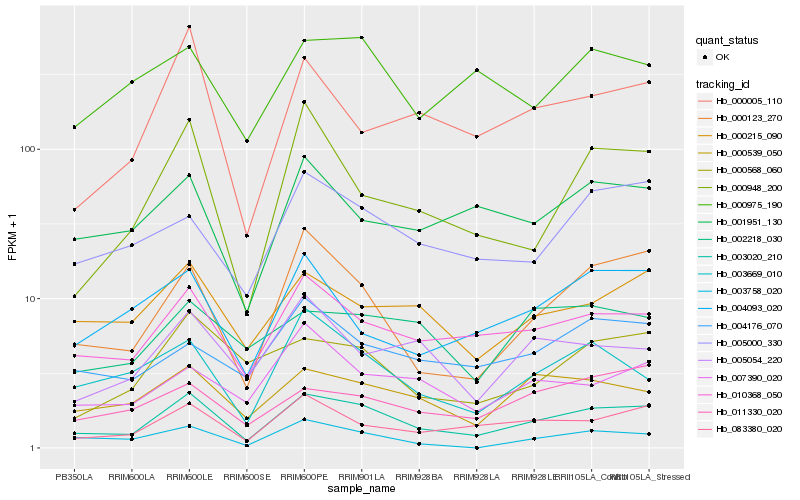
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_210 |
0.0 |
- |
- |
Structural maintenance of chromosomes 2-2 -like protein [Gossypium arboreum] |
| 2 |
Hb_010368_050 |
0.1215931805 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 3 |
Hb_000123_270 |
0.1354365529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_083380_020 |
0.1369546542 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 5 |
Hb_000539_050 |
0.145622712 |
- |
- |
bile acid:sodium symporter, putative [Ricinus communis] |
| 6 |
Hb_000215_090 |
0.1469472167 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_005000_330 |
0.1528944474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003669_010 |
0.1543661058 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic [Populus euphratica] |
| 9 |
Hb_001951_130 |
0.1600692634 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 10 |
Hb_011330_020 |
0.1635820554 |
transcription factor |
TF Family: ARID |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000568_060 |
0.1652081774 |
- |
- |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 12 |
Hb_004176_070 |
0.1654816341 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000005_110 |
0.1656180593 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 14 |
Hb_005054_220 |
0.167575077 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 15 |
Hb_004093_020 |
0.1675781555 |
- |
- |
Calcium-binding EF-hand family protein [Theobroma cacao] |
| 16 |
Hb_007390_020 |
0.1689278116 |
- |
- |
PREDICTED: protein FMP32, mitochondrial-like [Populus euphratica] |
| 17 |
Hb_000975_190 |
0.1710464272 |
- |
- |
histone H4 [Zea mays] |
| 18 |
Hb_003758_020 |
0.1713120754 |
- |
- |
UGTPg34 [Panax ginseng] |
| 19 |
Hb_000948_200 |
0.1722072002 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 20 |
Hb_002218_030 |
0.1735218706 |
- |
- |
hypothetical protein B456_007G345200 [Gossypium raimondii] |