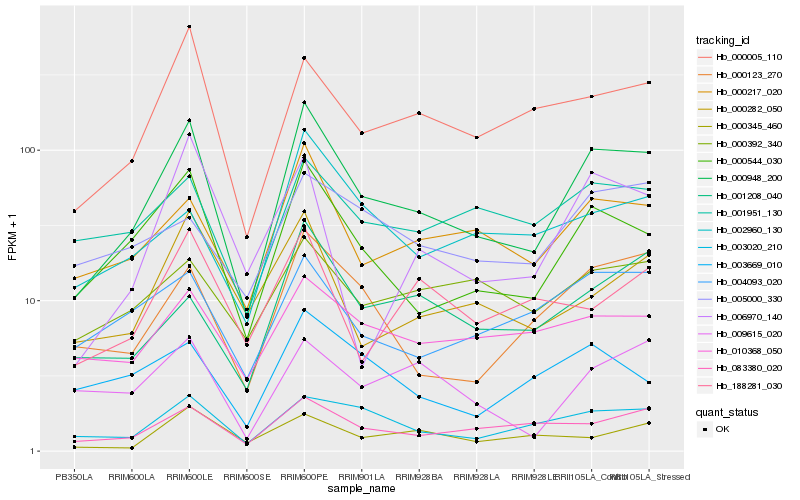
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000948_200 |
0.0 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 2 |
Hb_000544_030 |
0.1270933738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000217_020 |
0.1561758107 |
- |
- |
AMME syndrome candidateprotein 1 protein, putative [Ricinus communis] |
| 4 |
Hb_000123_270 |
0.164741455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000005_110 |
0.1682383806 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 6 |
Hb_003020_210 |
0.1722072002 |
- |
- |
Structural maintenance of chromosomes 2-2 -like protein [Gossypium arboreum] |
| 7 |
Hb_083380_020 |
0.1735219793 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 8 |
Hb_000282_050 |
0.1818527079 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 9 |
Hb_004093_020 |
0.1851575648 |
- |
- |
Calcium-binding EF-hand family protein [Theobroma cacao] |
| 10 |
Hb_001208_040 |
0.1885732792 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |
| 11 |
Hb_002960_130 |
0.1915630246 |
- |
- |
hypothetical protein B456_008G036100 [Gossypium raimondii] |
| 12 |
Hb_010368_050 |
0.1922046701 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 13 |
Hb_006970_140 |
0.1942744066 |
- |
- |
PREDICTED: histone H3.2-like [Solanum lycopersicum] |
| 14 |
Hb_001951_130 |
0.1952981358 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 15 |
Hb_009615_020 |
0.1984503375 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 7-like [Jatropha curcas] |
| 16 |
Hb_188281_030 |
0.2020298371 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 17 |
Hb_003669_010 |
0.2025317677 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic [Populus euphratica] |
| 18 |
Hb_000392_340 |
0.2026045123 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29 [Jatropha curcas] |
| 19 |
Hb_000345_460 |
0.2047376961 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 20 |
Hb_005000_330 |
0.2064272495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |