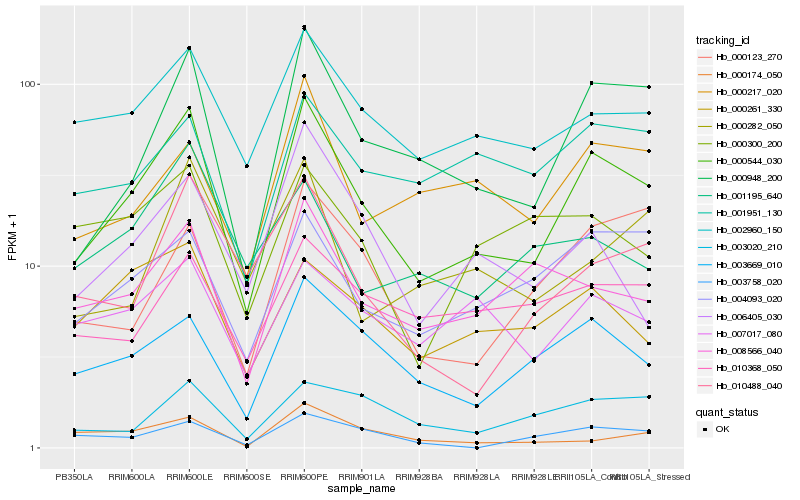
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000544_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000948_200 |
0.1270933738 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 3 |
Hb_003669_010 |
0.1572047162 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic [Populus euphratica] |
| 4 |
Hb_002960_150 |
0.1603850242 |
- |
- |
hypothetical protein CISIN_1g029215mg [Citrus sinensis] |
| 5 |
Hb_004093_020 |
0.1704417682 |
- |
- |
Calcium-binding EF-hand family protein [Theobroma cacao] |
| 6 |
Hb_008566_040 |
0.1766375056 |
- |
- |
PREDICTED: uncharacterized protein LOC105647590 isoform X1 [Jatropha curcas] |
| 7 |
Hb_010488_040 |
0.1811950428 |
- |
- |
PREDICTED: phosphomannomutase [Populus euphratica] |
| 8 |
Hb_000261_330 |
0.1815950767 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Amborella trichopoda] |
| 9 |
Hb_000217_020 |
0.1833879069 |
- |
- |
AMME syndrome candidateprotein 1 protein, putative [Ricinus communis] |
| 10 |
Hb_007017_080 |
0.1862513729 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_006405_030 |
0.1878546074 |
- |
- |
hypothetical protein VITISV_043238 [Vitis vinifera] |
| 12 |
Hb_003020_210 |
0.1898956006 |
- |
- |
Structural maintenance of chromosomes 2-2 -like protein [Gossypium arboreum] |
| 13 |
Hb_000123_270 |
0.1904213594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010368_050 |
0.193515748 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 15 |
Hb_000282_050 |
0.1939454142 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 16 |
Hb_001951_130 |
0.1946417713 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 17 |
Hb_003758_020 |
0.1980509747 |
- |
- |
UGTPg34 [Panax ginseng] |
| 18 |
Hb_000174_050 |
0.200758772 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 [Populus euphratica] |
| 19 |
Hb_000300_200 |
0.2018699027 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_001195_640 |
0.2020918605 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |