| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
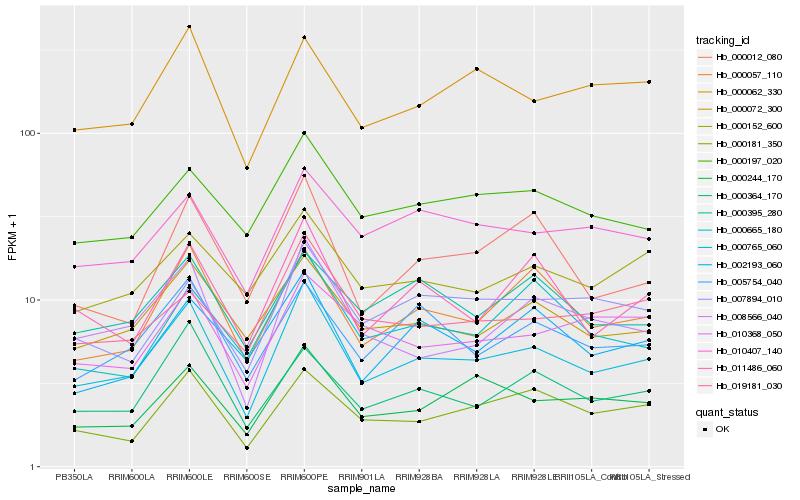
Hb_000765_060 |
0.0 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 2 |
Hb_008566_040 |
0.0981613011 |
- |
- |
PREDICTED: uncharacterized protein LOC105647590 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000057_110 |
0.1063013881 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 4 |
Hb_000072_300 |
0.1066689612 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 5 |
Hb_011486_060 |
0.1110208881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000244_170 |
0.111389477 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 7 |
Hb_000181_350 |
0.1135139009 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 8 |
Hb_005754_040 |
0.1172028593 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000197_020 |
0.1173280178 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 10 |
Hb_000012_080 |
0.1174482959 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 11 |
Hb_000364_170 |
0.1178565193 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 12 |
Hb_000062_330 |
0.1212557974 |
- |
- |
unknown [Lotus japonicus] |
| 13 |
Hb_010407_140 |
0.1235057404 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 14 |
Hb_007894_010 |
0.124302629 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 15 |
Hb_002193_060 |
0.1243825497 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 16 |
Hb_019181_030 |
0.1267853462 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 17 |
Hb_000152_600 |
0.126937965 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 18 |
Hb_000395_280 |
0.1296088379 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 19 |
Hb_010368_050 |
0.1308262829 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 20 |
Hb_000665_180 |
0.1309817413 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |