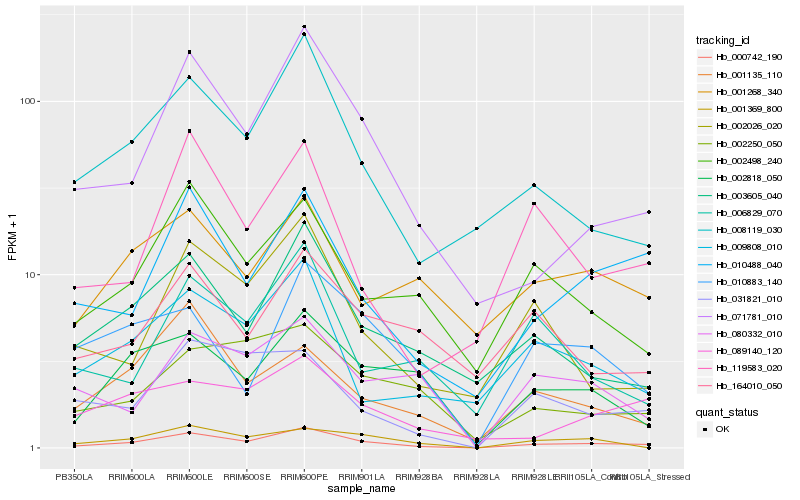
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000742_190 |
0.0 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_071781_010 |
0.1679417155 |
- |
- |
hypothetical protein POPTR_0010s13190g [Populus trichocarpa] |
| 3 |
Hb_010488_040 |
0.1685730267 |
- |
- |
PREDICTED: phosphomannomutase [Populus euphratica] |
| 4 |
Hb_008119_030 |
0.1699593848 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 5 |
Hb_003605_040 |
0.1769726304 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002818_050 |
0.1821799978 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 7 |
Hb_002498_240 |
0.189387662 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009808_010 |
0.1908705903 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_089140_120 |
0.192893688 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 10 |
Hb_002026_020 |
0.1976847696 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 11 |
Hb_031821_010 |
0.2009480242 |
- |
- |
ccd1, putative [Ricinus communis] |
| 12 |
Hb_002250_050 |
0.202117896 |
- |
- |
PREDICTED: uncharacterized protein LOC105638972 [Jatropha curcas] |
| 13 |
Hb_010883_140 |
0.2043599288 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_119583_020 |
0.2046771091 |
- |
- |
PREDICTED: uncharacterized protein LOC105628090 [Jatropha curcas] |
| 15 |
Hb_001268_340 |
0.2057891369 |
- |
- |
Actin, putative [Ricinus communis] |
| 16 |
Hb_080332_010 |
0.2068015238 |
- |
- |
hypothetical protein POPTR_0018s10590g [Populus trichocarpa] |
| 17 |
Hb_001135_110 |
0.2088901801 |
- |
- |
- |
| 18 |
Hb_001369_800 |
0.209783521 |
- |
- |
hypothetical protein B456_011G005000 [Gossypium raimondii] |
| 19 |
Hb_164010_050 |
0.211016194 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 20 |
Hb_006829_070 |
0.2134177909 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |