| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
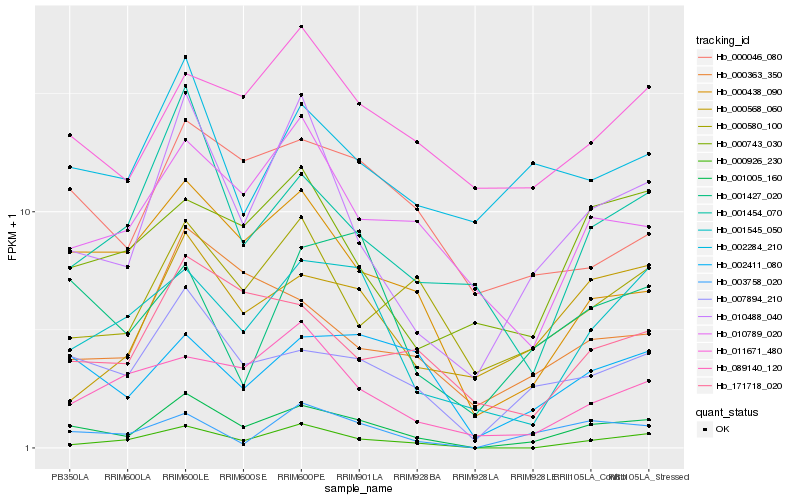
Hb_001005_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007894_210 |
0.143524845 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 3 |
Hb_010488_040 |
0.1553195764 |
- |
- |
PREDICTED: phosphomannomutase [Populus euphratica] |
| 4 |
Hb_000438_090 |
0.1671043046 |
- |
- |
PREDICTED: mini-chromosome maintenance complex-binding protein [Jatropha curcas] |
| 5 |
Hb_000926_230 |
0.1823672393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001545_050 |
0.1849012536 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 7 |
Hb_002411_080 |
0.1860922156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003758_020 |
0.1931888073 |
- |
- |
UGTPg34 [Panax ginseng] |
| 9 |
Hb_171718_020 |
0.196152764 |
- |
- |
hypothetical protein JCGZ_00274 [Jatropha curcas] |
| 10 |
Hb_010789_020 |
0.1999485279 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 11 |
Hb_001454_070 |
0.2000992121 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 12 |
Hb_000363_350 |
0.2041370994 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like [Fragaria vesca subsp. vesca] |
| 13 |
Hb_000580_100 |
0.2043623744 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000046_080 |
0.2064892532 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL28 [Jatropha curcas] |
| 15 |
Hb_089140_120 |
0.2073738133 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 16 |
Hb_011671_480 |
0.2077010376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000568_060 |
0.2092253726 |
- |
- |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 18 |
Hb_000743_030 |
0.2097064218 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 19 |
Hb_001427_020 |
0.2111787032 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 20 |
Hb_002284_210 |
0.2146877393 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |