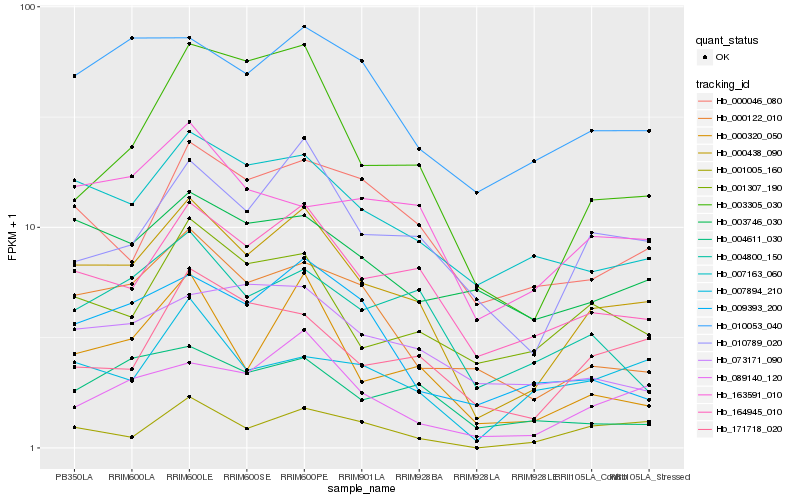
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000438_090 |
0.0 |
- |
- |
PREDICTED: mini-chromosome maintenance complex-binding protein [Jatropha curcas] |
| 2 |
Hb_164945_010 |
0.1167804494 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 3 |
Hb_010789_020 |
0.1372662862 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 4 |
Hb_089140_120 |
0.1410681364 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 5 |
Hb_007163_060 |
0.1417698617 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 6 |
Hb_000320_050 |
0.1449055802 |
- |
- |
bcr-associated protein, bap, putative [Ricinus communis] |
| 7 |
Hb_000122_010 |
0.1460576746 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_010053_040 |
0.1500655242 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 9 |
Hb_003305_030 |
0.1501717773 |
- |
- |
- |
| 10 |
Hb_001307_190 |
0.1521248103 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000046_080 |
0.1533421484 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL28 [Jatropha curcas] |
| 12 |
Hb_171718_020 |
0.1554705311 |
- |
- |
hypothetical protein JCGZ_00274 [Jatropha curcas] |
| 13 |
Hb_163591_010 |
0.1566058654 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Populus euphratica] |
| 14 |
Hb_004800_150 |
0.1583483459 |
transcription factor |
TF Family: SNF2 |
PREDICTED: ATP-dependent DNA helicase DDM1 [Jatropha curcas] |
| 15 |
Hb_004611_030 |
0.1588790288 |
- |
- |
structural maintenance of chromosomes 5 smc5, putative [Ricinus communis] |
| 16 |
Hb_009393_200 |
0.160813734 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 17 |
Hb_073171_090 |
0.1648432554 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 18 |
Hb_007894_210 |
0.1663536055 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 19 |
Hb_001005_160 |
0.1671043046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003746_030 |
0.1674734456 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |