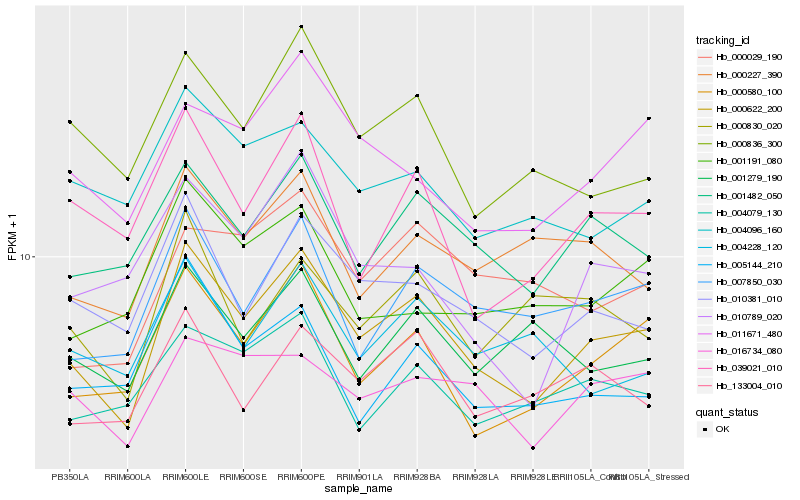
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000622_200 |
0.0 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 2 |
Hb_000580_100 |
0.1069066003 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 3 |
Hb_010381_010 |
0.1118873405 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_001482_050 |
0.1143605408 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 5 |
Hb_007850_030 |
0.1164419228 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 6 |
Hb_016734_080 |
0.1265548517 |
- |
- |
PREDICTED: pyridine nucleotide-disulfide oxidoreductase domain-containing protein 2 [Jatropha curcas] |
| 7 |
Hb_039021_010 |
0.1286332378 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 8 |
Hb_133004_010 |
0.1289071624 |
- |
- |
PREDICTED: cyclin-D3-3 [Jatropha curcas] |
| 9 |
Hb_000836_300 |
0.1305822638 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 10 |
Hb_004079_130 |
0.1324462139 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 11 |
Hb_010789_020 |
0.1344934278 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 12 |
Hb_011671_480 |
0.1350810765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000227_390 |
0.1358726714 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004096_160 |
0.1377393489 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 15 |
Hb_001279_190 |
0.1382453582 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000029_190 |
0.14131273 |
- |
- |
hypothetical protein CISIN_1g025338mg [Citrus sinensis] |
| 17 |
Hb_005144_210 |
0.1413644836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000830_020 |
0.141929846 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 19 |
Hb_001191_080 |
0.14237244 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_004228_120 |
0.1437834492 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |