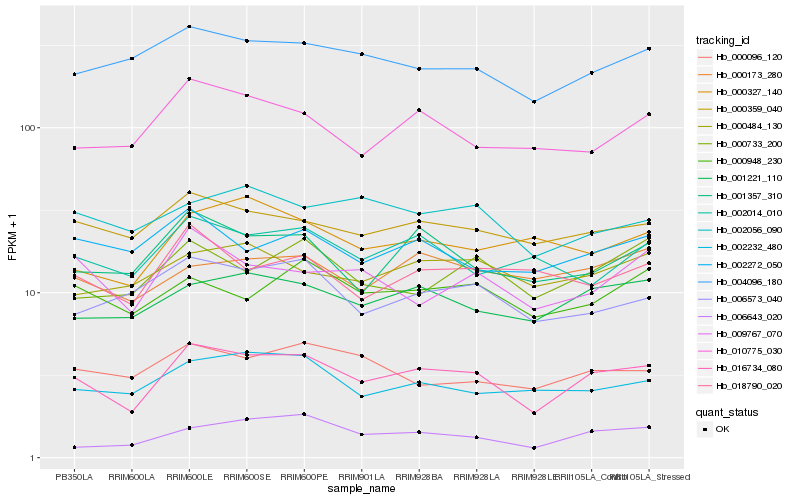
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016734_080 |
0.0 |
- |
- |
PREDICTED: pyridine nucleotide-disulfide oxidoreductase domain-containing protein 2 [Jatropha curcas] |
| 2 |
Hb_001221_110 |
0.1071406015 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 3 |
Hb_000359_040 |
0.1102509081 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001357_310 |
0.1110605011 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_010775_030 |
0.1138360585 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 6 |
Hb_004096_180 |
0.116032737 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 7 |
Hb_002232_480 |
0.1166385298 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 8 |
Hb_000948_230 |
0.1210120698 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_006573_040 |
0.1217983332 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_009767_070 |
0.1230223506 |
- |
- |
PREDICTED: uncharacterized protein LOC105631980 [Jatropha curcas] |
| 11 |
Hb_018790_020 |
0.1232672807 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000484_130 |
0.1240305768 |
- |
- |
splicing factor, putative [Ricinus communis] |
| 13 |
Hb_002056_090 |
0.124352781 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 14 |
Hb_000733_200 |
0.1244880249 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 15 |
Hb_000096_120 |
0.1245437679 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 16 |
Hb_002014_010 |
0.1246091237 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_000173_280 |
0.1253055024 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 18 |
Hb_006643_020 |
0.125606822 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 19 |
Hb_000327_140 |
0.1256246603 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002272_050 |
0.1256469246 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |