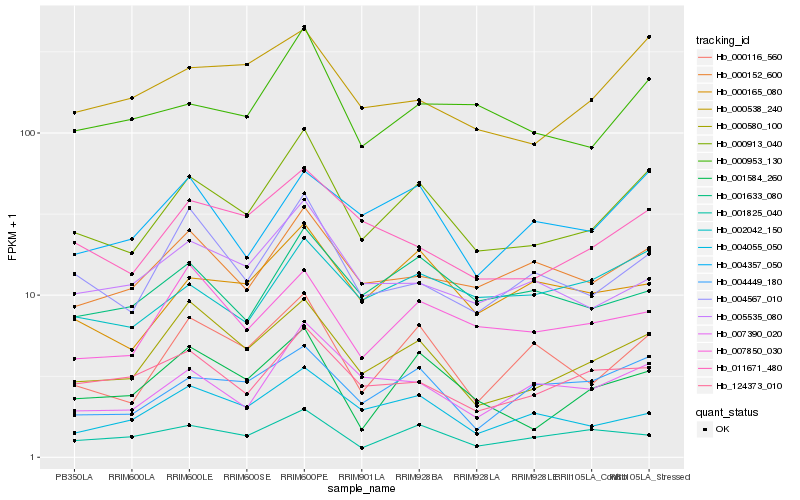
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000913_040 |
0.0 |
- |
- |
Methyltransferase-related protein, putative [Theobroma cacao] |
| 2 |
Hb_000580_100 |
0.1077002631 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000953_130 |
0.1176246632 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 4 |
Hb_000116_560 |
0.1196290866 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 5 |
Hb_007390_020 |
0.124383885 |
- |
- |
PREDICTED: protein FMP32, mitochondrial-like [Populus euphratica] |
| 6 |
Hb_011671_480 |
0.1263152497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001584_260 |
0.1270869641 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_004449_180 |
0.1279995965 |
- |
- |
PREDICTED: SAC3 family protein 1 [Jatropha curcas] |
| 9 |
Hb_000538_240 |
0.1291216068 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 10 |
Hb_000152_600 |
0.1292169501 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 11 |
Hb_000165_080 |
0.1301314328 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004567_010 |
0.1305865453 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 13 |
Hb_001633_080 |
0.1356868246 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 14 |
Hb_004055_050 |
0.1361956927 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 15 |
Hb_124373_010 |
0.136373111 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 16 |
Hb_002042_150 |
0.1372332071 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 17 |
Hb_005535_080 |
0.1373514588 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 18 |
Hb_004357_050 |
0.1375552827 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 19 |
Hb_001825_040 |
0.1390444148 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 20 |
Hb_007850_030 |
0.1397722859 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |