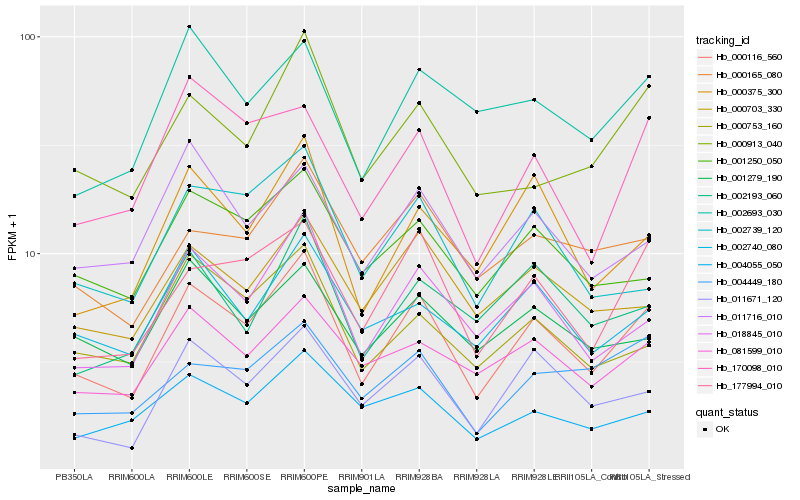
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_560 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 2 |
Hb_018845_010 |
0.0956075647 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 3 |
Hb_177994_010 |
0.103244534 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 4 |
Hb_011671_120 |
0.1116315426 |
- |
- |
hypothetical protein JCGZ_26275 [Jatropha curcas] |
| 5 |
Hb_000375_300 |
0.1119259735 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 6 |
Hb_081599_010 |
0.1160579423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000913_040 |
0.1196290866 |
- |
- |
Methyltransferase-related protein, putative [Theobroma cacao] |
| 8 |
Hb_002740_080 |
0.1206009232 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 9 |
Hb_000703_330 |
0.1227715428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000753_160 |
0.123531953 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 11 |
Hb_001250_050 |
0.1246844637 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 12 |
Hb_170098_010 |
0.1248620007 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 13 |
Hb_000165_080 |
0.1252960324 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004055_050 |
0.1261605249 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 15 |
Hb_002193_060 |
0.1274640378 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 16 |
Hb_011716_010 |
0.127492444 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 17 |
Hb_002693_030 |
0.127516191 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 18 |
Hb_004449_180 |
0.1288971823 |
- |
- |
PREDICTED: SAC3 family protein 1 [Jatropha curcas] |
| 19 |
Hb_002739_120 |
0.1298961719 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 20 |
Hb_001279_190 |
0.1302332327 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |