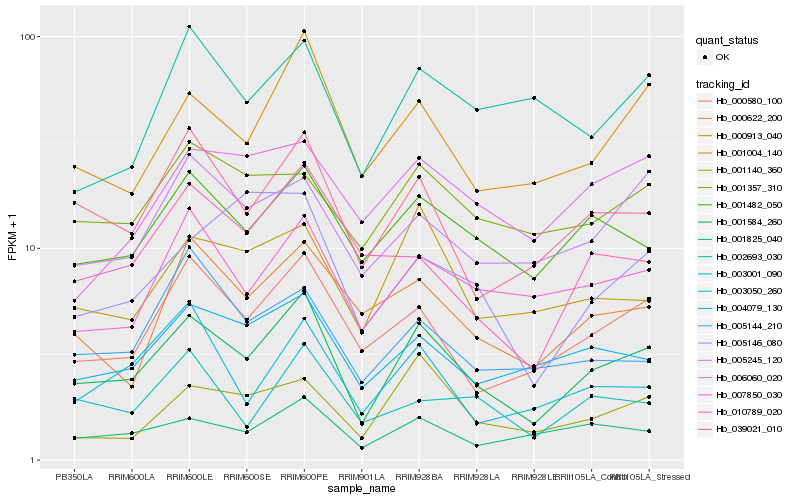
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_260 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_039021_010 |
0.1255299441 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 3 |
Hb_000913_040 |
0.1270869641 |
- |
- |
Methyltransferase-related protein, putative [Theobroma cacao] |
| 4 |
Hb_000580_100 |
0.1288887985 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001825_040 |
0.136689302 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 6 |
Hb_001482_050 |
0.1367237462 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 7 |
Hb_007850_030 |
0.1422527422 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 8 |
Hb_004079_130 |
0.1430427709 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 9 |
Hb_001004_140 |
0.147200862 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 10 |
Hb_000622_200 |
0.1535781364 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 11 |
Hb_005245_120 |
0.155165459 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 12 |
Hb_001140_360 |
0.1557711027 |
- |
- |
- |
| 13 |
Hb_003050_260 |
0.1604494874 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 14 |
Hb_010789_020 |
0.1606559659 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 15 |
Hb_003001_090 |
0.1614105791 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 16 |
Hb_001357_310 |
0.1635439095 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_005144_210 |
0.1674565671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005146_080 |
0.1705301479 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002693_030 |
0.1706406638 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 20 |
Hb_006060_020 |
0.1714747426 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |