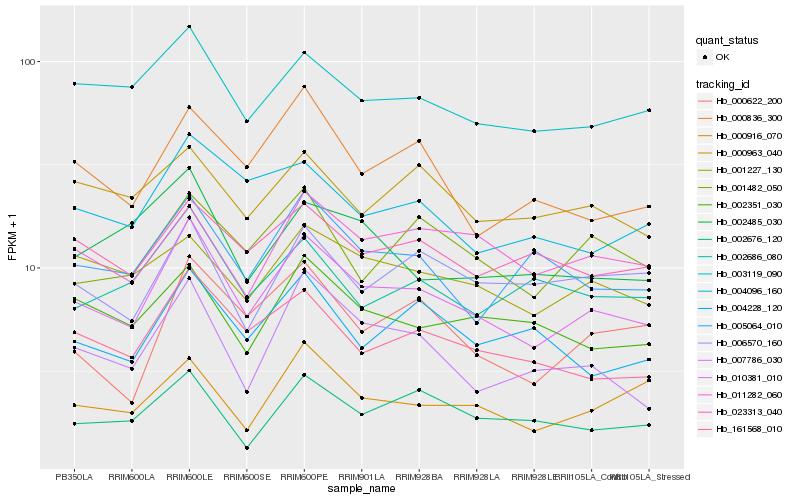
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010381_010 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_161568_010 |
0.0896327021 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_003119_090 |
0.0988011136 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 4 |
Hb_000836_300 |
0.1081932296 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 5 |
Hb_007786_030 |
0.1097752415 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 6 |
Hb_004096_160 |
0.1105342756 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 7 |
Hb_000622_200 |
0.1118873405 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 8 |
Hb_001227_130 |
0.1120077448 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 9 |
Hb_005064_010 |
0.1125971608 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 10 |
Hb_002351_030 |
0.1126863219 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_011282_060 |
0.1127337116 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 12 |
Hb_023313_040 |
0.1128924742 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001482_050 |
0.1137240432 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 14 |
Hb_004228_120 |
0.1161164324 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 15 |
Hb_002676_120 |
0.1170927324 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 16 |
Hb_002686_080 |
0.1187701071 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein ING2-like [Jatropha curcas] |
| 17 |
Hb_006570_160 |
0.1195464272 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 18 |
Hb_000916_070 |
0.1200603139 |
- |
- |
PREDICTED: sister chromatid cohesion protein DCC1-like [Jatropha curcas] |
| 19 |
Hb_002485_030 |
0.1204245802 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 20 |
Hb_000963_040 |
0.121344168 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |