| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
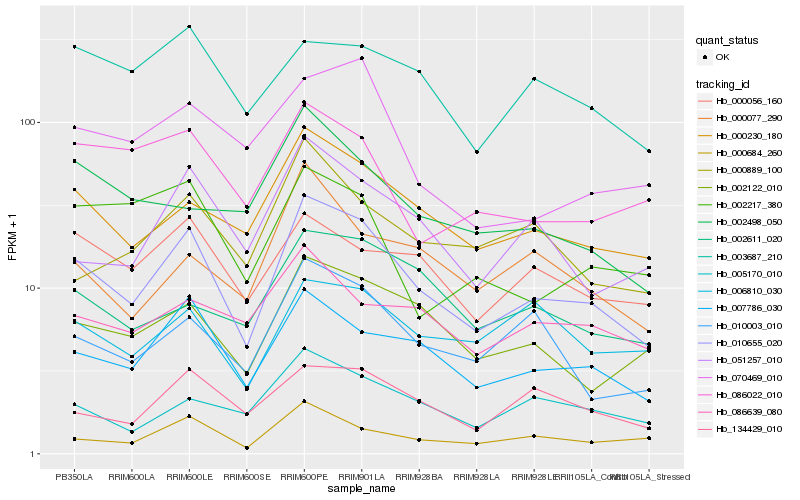
| 1 |
Hb_010655_020 |
0.0 |
- |
- |
hypothetical protein CICLE_v10032410mg [Citrus clementina] |
| 2 |
Hb_007786_030 |
0.1151125854 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 3 |
Hb_000230_180 |
0.1249437281 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 4 |
Hb_002122_010 |
0.1361319547 |
- |
- |
- |
| 5 |
Hb_000684_260 |
0.1387398159 |
- |
- |
PREDICTED: probable DNA helicase MCM8 isoform X2 [Jatropha curcas] |
| 6 |
Hb_010003_010 |
0.1435871842 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 7 |
Hb_051257_010 |
0.146202931 |
- |
- |
- |
| 8 |
Hb_134429_010 |
0.1512589073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005170_010 |
0.152227334 |
- |
- |
- |
| 10 |
Hb_000056_160 |
0.158540857 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP23 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002611_020 |
0.1633446914 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 12 |
Hb_086022_010 |
0.1652784203 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 13 |
Hb_000077_290 |
0.1654656988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000889_100 |
0.1663856488 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 15 |
Hb_006810_030 |
0.1664413776 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_070469_010 |
0.1677074151 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 17 |
Hb_003687_210 |
0.1689708063 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 18 |
Hb_086639_080 |
0.1717589108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002498_050 |
0.1729320245 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 20 |
Hb_002217_380 |
0.1731850804 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |