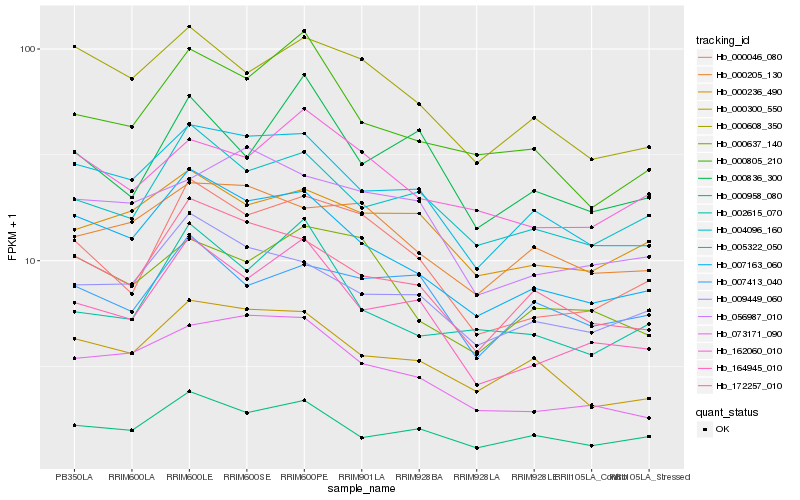
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_080 |
0.0 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL28 [Jatropha curcas] |
| 2 |
Hb_164945_010 |
0.0927563506 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 3 |
Hb_007163_060 |
0.1031325117 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 4 |
Hb_004096_160 |
0.1087900401 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 5 |
Hb_172257_010 |
0.1135985107 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 6 |
Hb_000608_350 |
0.1148132922 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 7 |
Hb_162060_010 |
0.1157536037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000637_140 |
0.1170277961 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 9 |
Hb_009449_060 |
0.1175221005 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 10 |
Hb_005322_050 |
0.1181947494 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 11 |
Hb_007413_040 |
0.1207723972 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 12 |
Hb_000958_080 |
0.1245518684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000236_490 |
0.1249020749 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 14 |
Hb_000805_210 |
0.126276981 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 15 |
Hb_000836_300 |
0.1281318982 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 16 |
Hb_056987_010 |
0.1303259533 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 17 |
Hb_000300_550 |
0.1305374569 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 18 |
Hb_000205_130 |
0.1309050973 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_002615_070 |
0.1323024101 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 20 |
Hb_073171_090 |
0.1342844335 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |