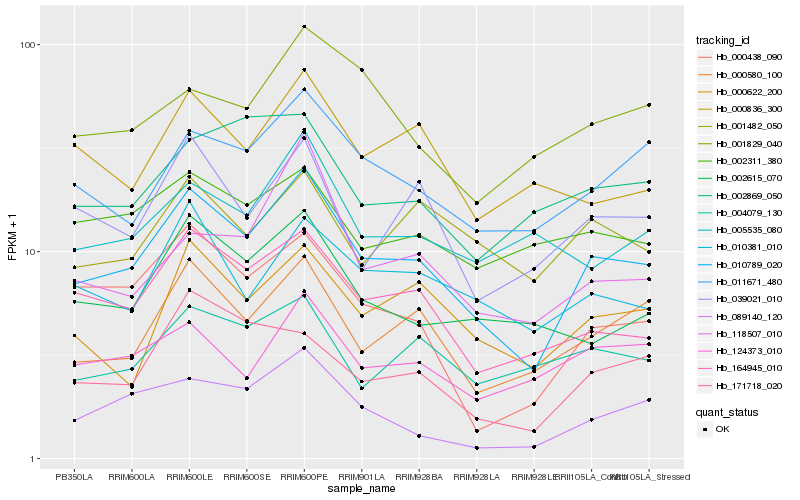
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010789_020 |
0.0 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 2 |
Hb_164945_010 |
0.1122789512 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 3 |
Hb_000580_100 |
0.118579523 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 4 |
Hb_011671_480 |
0.1247087963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010381_010 |
0.1334919278 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_039021_010 |
0.1335341778 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 7 |
Hb_000622_200 |
0.1344934278 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 8 |
Hb_002615_070 |
0.1361432476 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000438_090 |
0.1372662862 |
- |
- |
PREDICTED: mini-chromosome maintenance complex-binding protein [Jatropha curcas] |
| 10 |
Hb_124373_010 |
0.138681611 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 11 |
Hb_089140_120 |
0.1405236354 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 12 |
Hb_171718_020 |
0.143554273 |
- |
- |
hypothetical protein JCGZ_00274 [Jatropha curcas] |
| 13 |
Hb_004079_130 |
0.143796698 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 14 |
Hb_000836_300 |
0.1459229932 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 15 |
Hb_001482_050 |
0.1459356131 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 16 |
Hb_118507_010 |
0.1466018553 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 17 |
Hb_002311_380 |
0.1485239584 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 18 |
Hb_001829_040 |
0.1490436679 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 19 |
Hb_005535_080 |
0.1494659331 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 20 |
Hb_002869_050 |
0.1504490526 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |