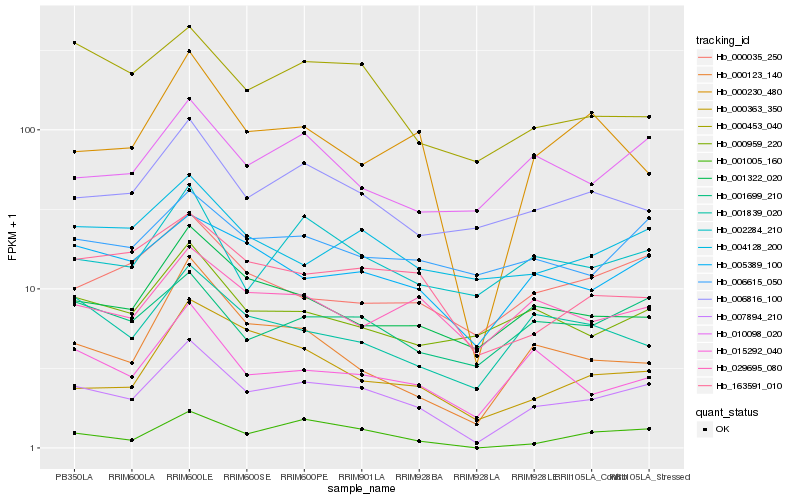
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_210 |
0.0 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 2 |
Hb_005389_100 |
0.1325099398 |
- |
- |
PREDICTED: methyltransferase-like protein 5 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001699_210 |
0.1408038629 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001005_160 |
0.143524845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001322_020 |
0.1465353019 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 6 |
Hb_015292_040 |
0.147627176 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 7 |
Hb_163591_010 |
0.1486672107 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Populus euphratica] |
| 8 |
Hb_006816_100 |
0.1488582285 |
- |
- |
hypothetical protein CISIN_1g0373181mg, partial [Citrus sinensis] |
| 9 |
Hb_002284_210 |
0.1495609053 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |
| 10 |
Hb_004128_200 |
0.1512792634 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 11 |
Hb_000230_480 |
0.1513722037 |
- |
- |
PREDICTED: endo-1,3;1,4-beta-D-glucanase-like [Jatropha curcas] |
| 12 |
Hb_000035_250 |
0.1522666584 |
- |
- |
PREDICTED: uncharacterized protein LOC105637544 [Jatropha curcas] |
| 13 |
Hb_001839_020 |
0.1537169497 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 14 |
Hb_010098_020 |
0.155741717 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_000123_140 |
0.1567968891 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 16 |
Hb_029695_080 |
0.1573648567 |
- |
- |
PREDICTED: NAD-dependent protein deacetylase SRT1 [Prunus mume] |
| 17 |
Hb_006615_050 |
0.1578091722 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 18 |
Hb_000453_040 |
0.1587934291 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 19 |
Hb_000959_220 |
0.1590075598 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000363_350 |
0.1595307895 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like [Fragaria vesca subsp. vesca] |