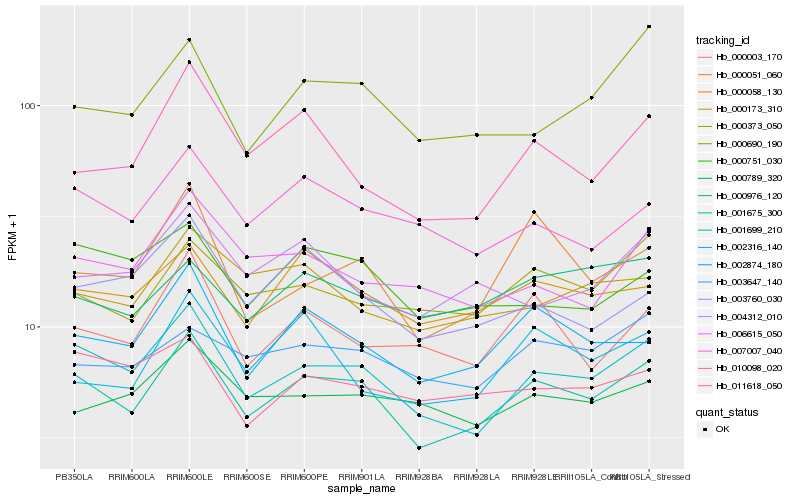
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001675_300 |
0.0 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001699_210 |
0.0595652716 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002874_180 |
0.0886934767 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000058_130 |
0.0951113109 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 5 |
Hb_000373_050 |
0.0983373858 |
- |
- |
PREDICTED: protein WHI3 [Jatropha curcas] |
| 6 |
Hb_007007_040 |
0.1004995881 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 7 |
Hb_000003_170 |
0.1017012028 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 8 |
Hb_000051_060 |
0.1040987072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003647_140 |
0.1059152856 |
- |
- |
PREDICTED: uncharacterized protein LOC105646673 [Jatropha curcas] |
| 10 |
Hb_000976_120 |
0.1069687713 |
- |
- |
PREDICTED: DNA polymerase zeta processivity subunit [Jatropha curcas] |
| 11 |
Hb_010098_020 |
0.1071670008 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_000690_190 |
0.107542172 |
- |
- |
40S ribosomal protein S7, putative [Ricinus communis] |
| 13 |
Hb_000751_030 |
0.1083078708 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 14 |
Hb_000173_310 |
0.1088895796 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006615_050 |
0.109138013 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 16 |
Hb_011618_050 |
0.1099353659 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 17 |
Hb_002316_140 |
0.1112899499 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |
| 18 |
Hb_004312_010 |
0.1121086172 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003760_030 |
0.1124335607 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 20 |
Hb_000789_320 |
0.1130334678 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |