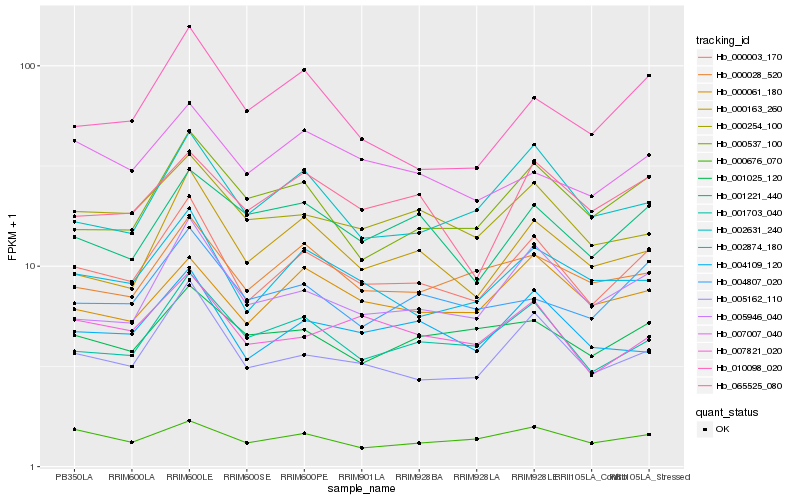
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_170 |
0.0 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 2 |
Hb_005162_110 |
0.0688477827 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 3 |
Hb_002874_180 |
0.0762347973 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001703_040 |
0.0793027045 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005946_040 |
0.0798342271 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000163_260 |
0.0844209688 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 7 |
Hb_004109_120 |
0.0903230272 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000028_520 |
0.0926954228 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 9 |
Hb_000254_100 |
0.093249757 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 10 |
Hb_004807_020 |
0.0934084434 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 11 |
Hb_007821_020 |
0.0935450567 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000537_100 |
0.0943117072 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000676_070 |
0.0944744025 |
- |
- |
PREDICTED: uncharacterized protein LOC105634617 [Jatropha curcas] |
| 14 |
Hb_000061_180 |
0.0959405728 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 15 |
Hb_002631_240 |
0.096164468 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 16 |
Hb_065525_080 |
0.0989266303 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 14 [Populus euphratica] |
| 17 |
Hb_010098_020 |
0.0994640118 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 18 |
Hb_001221_440 |
0.0996372378 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001025_120 |
0.0996813116 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_007007_040 |
0.1002294855 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |