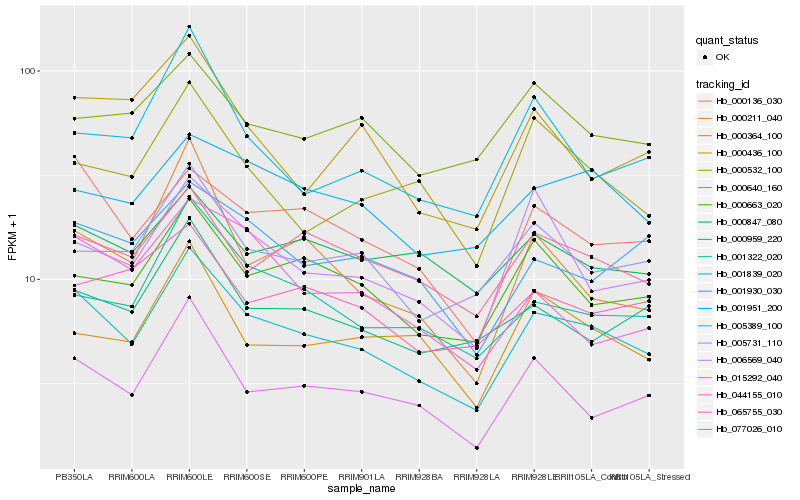
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001839_020 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 2 |
Hb_000136_030 |
0.1009549165 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 3 |
Hb_015292_040 |
0.1034893447 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 4 |
Hb_000663_020 |
0.1086803066 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_001322_020 |
0.1088712783 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 6 |
Hb_077026_010 |
0.1103528436 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 7 |
Hb_005731_110 |
0.112099633 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 8 |
Hb_000959_220 |
0.1133874918 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_001951_200 |
0.1166888825 |
- |
- |
PREDICTED: cold-regulated 413 inner membrane protein 1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_005389_100 |
0.1186632291 |
- |
- |
PREDICTED: methyltransferase-like protein 5 isoform X2 [Jatropha curcas] |
| 11 |
Hb_006569_040 |
0.1206189397 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 12 |
Hb_065755_030 |
0.1227591952 |
- |
- |
- |
| 13 |
Hb_000211_040 |
0.1251151759 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 14 |
Hb_000364_100 |
0.1252372394 |
- |
- |
PREDICTED: uncharacterized protein LOC105639739 [Jatropha curcas] |
| 15 |
Hb_000847_080 |
0.1261698938 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 16 |
Hb_000532_100 |
0.1274074535 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 17 |
Hb_001930_030 |
0.1283228496 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000436_100 |
0.1283590886 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 19 |
Hb_044155_010 |
0.1292713759 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000640_160 |
0.1301886368 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |