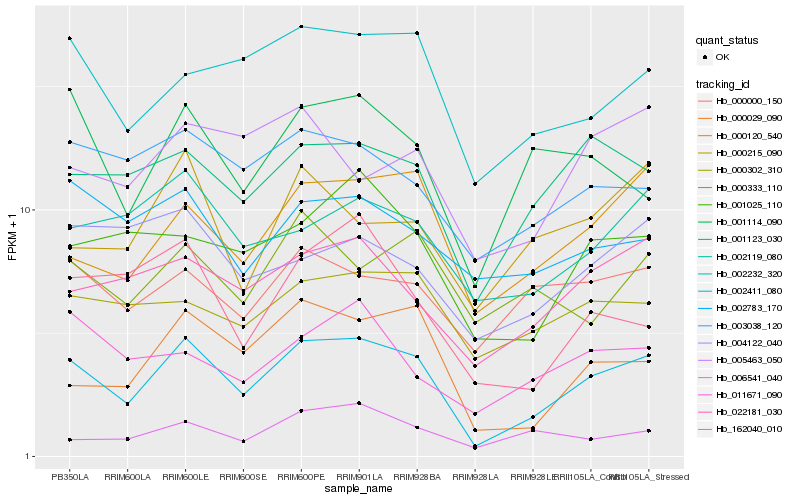
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002411_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000120_540 |
0.1374610593 |
- |
- |
translation initiation factor, putative [Ricinus communis] |
| 3 |
Hb_000029_090 |
0.1444419947 |
- |
- |
Nicalin precursor, putative [Ricinus communis] |
| 4 |
Hb_002232_320 |
0.1447499207 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_001123_030 |
0.145048709 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_022181_030 |
0.1490339572 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001025_110 |
0.1519109482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001114_090 |
0.1576823037 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 9 |
Hb_002119_080 |
0.1597180118 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 10 |
Hb_003038_120 |
0.1609557219 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 11 |
Hb_000000_150 |
0.1614362633 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 12 |
Hb_000302_310 |
0.1619964722 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 13 |
Hb_162040_010 |
0.1667411296 |
- |
- |
- |
| 14 |
Hb_011671_090 |
0.1677238404 |
- |
- |
PREDICTED: protein RFT1 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_005463_050 |
0.1681007205 |
- |
- |
PREDICTED: uncharacterized protein LOC105632514 [Jatropha curcas] |
| 16 |
Hb_000215_090 |
0.1682519287 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 17 |
Hb_006541_040 |
0.1707915634 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 2 [Jatropha curcas] |
| 18 |
Hb_002783_170 |
0.1713491816 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 19 |
Hb_004122_040 |
0.1718272254 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 20 |
Hb_000333_110 |
0.1725620767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |