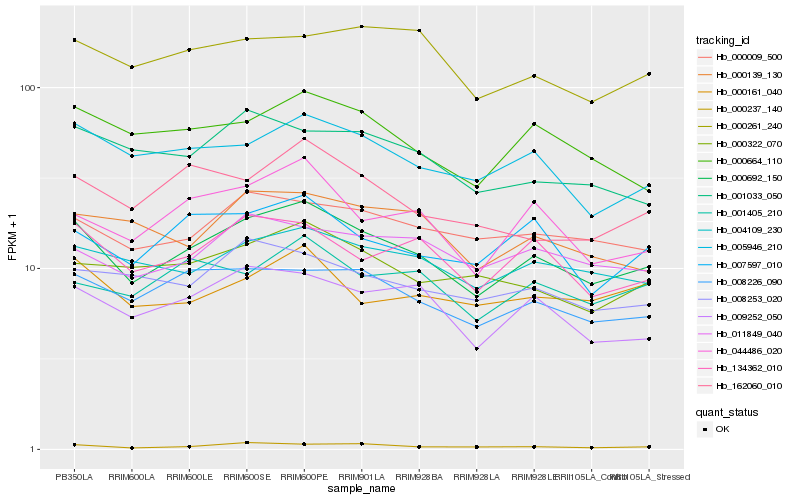
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000692_150 |
0.0 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 3 [Jatropha curcas] |
| 2 |
Hb_009252_050 |
0.0836208131 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_008226_090 |
0.0842309479 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005946_210 |
0.0865259819 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 5 |
Hb_134362_010 |
0.0878042111 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 6 |
Hb_004109_230 |
0.0883979036 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_044486_020 |
0.0917989524 |
- |
- |
CASTOR protein [Glycine max] |
| 8 |
Hb_000664_110 |
0.0922823528 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 9 |
Hb_000009_500 |
0.0936518287 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 10 |
Hb_007597_010 |
0.0966815684 |
- |
- |
hypothetical protein B456_010G072300 [Gossypium raimondii] |
| 11 |
Hb_000322_070 |
0.0983128956 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_162060_010 |
0.0989078874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008253_020 |
0.0992201964 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 14 |
Hb_011849_040 |
0.0992341288 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_000139_130 |
0.0996656372 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 16 |
Hb_001405_210 |
0.0998360697 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 17 |
Hb_000237_140 |
0.1010497829 |
- |
- |
PREDICTED: RNA-binding protein 28 [Jatropha curcas] |
| 18 |
Hb_001033_050 |
0.1031428265 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 19 |
Hb_000161_040 |
0.1031643029 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 20 |
Hb_000261_240 |
0.1032878232 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |