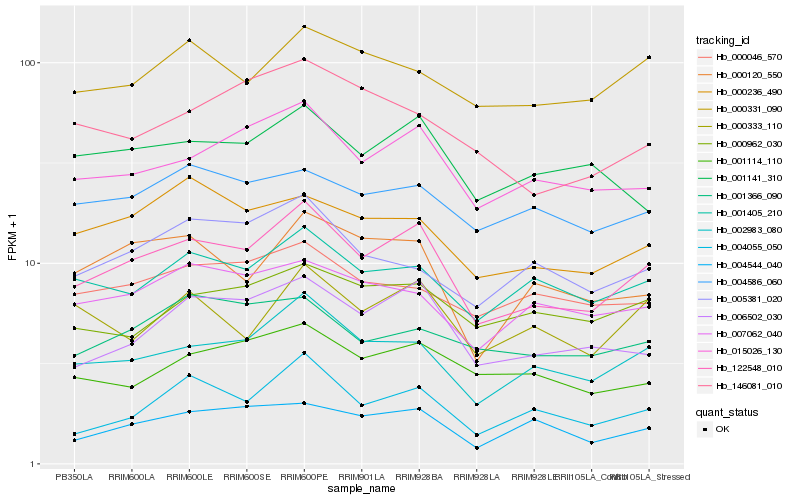
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_122548_010 |
0.0 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 2 |
Hb_002983_080 |
0.07765524 |
- |
- |
acetylornithine aminotransferase, partial [Prunus persica] |
| 3 |
Hb_015026_130 |
0.085887002 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 4 |
Hb_006502_030 |
0.0891007094 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 5 |
Hb_001405_210 |
0.1009514644 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 6 |
Hb_000120_550 |
0.1059587028 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 7 |
Hb_004055_050 |
0.1059675304 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 8 |
Hb_000236_490 |
0.1061775574 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 9 |
Hb_004544_040 |
0.1090262014 |
- |
- |
hypothetical protein PHAVU_010G103000g [Phaseolus vulgaris] |
| 10 |
Hb_001114_110 |
0.1108431949 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 11 |
Hb_000333_110 |
0.1109363513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001141_310 |
0.1109577617 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 13 |
Hb_007062_040 |
0.1113878064 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 14 |
Hb_000331_090 |
0.1118810983 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 15 |
Hb_004586_060 |
0.1138682648 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 16 |
Hb_005381_020 |
0.115147661 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 17 |
Hb_000962_030 |
0.1161155359 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 18 |
Hb_000046_570 |
0.1163237322 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 19 |
Hb_001366_090 |
0.1168865289 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1341380 [Ricinus communis] |
| 20 |
Hb_146081_010 |
0.1172612468 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |