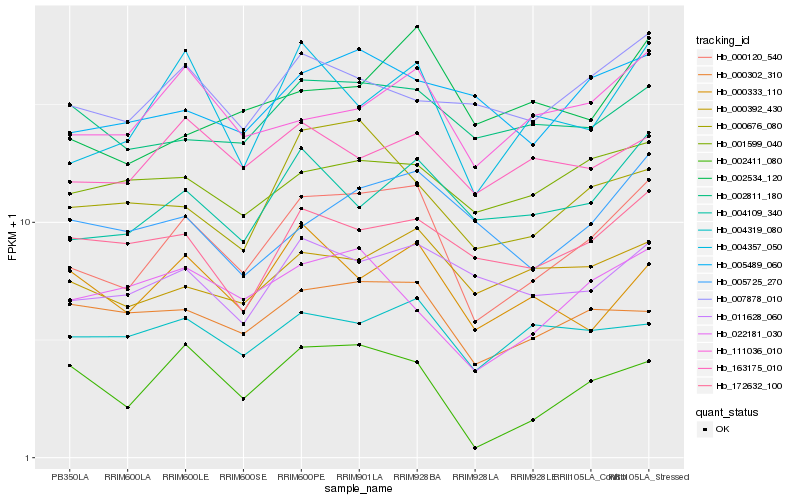
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_540 |
0.0 |
- |
- |
translation initiation factor, putative [Ricinus communis] |
| 2 |
Hb_004357_050 |
0.1118474498 |
- |
- |
PREDICTED: histone deacetylase HDT1-like [Jatropha curcas] |
| 3 |
Hb_004109_340 |
0.1180097076 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14-like [Jatropha curcas] |
| 4 |
Hb_111036_010 |
0.1262337661 |
- |
- |
Eukaryotic translation initiation factor 6-2 [Glycine soja] |
| 5 |
Hb_000392_430 |
0.129571844 |
- |
- |
PREDICTED: uncharacterized protein LOC105640501 [Jatropha curcas] |
| 6 |
Hb_011628_060 |
0.1299578718 |
- |
- |
Adenosine 3'-phospho 5'-phosphosulfate transporter, putative [Ricinus communis] |
| 7 |
Hb_000302_310 |
0.1318801284 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 8 |
Hb_000676_080 |
0.1338320248 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 9 |
Hb_172632_100 |
0.1364931337 |
- |
- |
PREDICTED: reticulon-like protein B22 [Jatropha curcas] |
| 10 |
Hb_005489_060 |
0.1369105938 |
- |
- |
Anamorsin, putative [Ricinus communis] |
| 11 |
Hb_002411_080 |
0.1374610593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004319_080 |
0.1382447012 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 13 |
Hb_002811_180 |
0.1383282925 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_022181_030 |
0.1390697186 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000333_110 |
0.1392640672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005725_270 |
0.1393893868 |
- |
- |
PREDICTED: uncharacterized protein LOC105630802 isoform X1 [Jatropha curcas] |
| 17 |
Hb_163175_010 |
0.1395637737 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 18 |
Hb_001599_040 |
0.1401304461 |
- |
- |
PREDICTED: suppressor protein SRP40 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007878_010 |
0.1401431661 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 4 [Jatropha curcas] |
| 20 |
Hb_002534_120 |
0.1401795478 |
- |
- |
PREDICTED: protein DCL, chloroplastic [Jatropha curcas] |