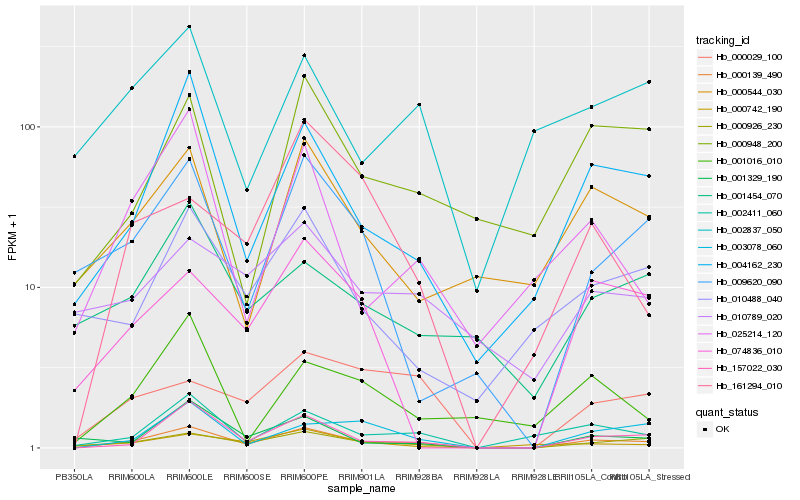
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_490 |
0.0 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 2 |
Hb_157022_030 |
0.1723691185 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 3 |
Hb_000926_230 |
0.1729643024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004162_230 |
0.2081191451 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 5 |
Hb_002411_060 |
0.2122700115 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 6 |
Hb_003078_060 |
0.2192959841 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 7 |
Hb_000029_100 |
0.2417907756 |
- |
- |
PREDICTED: nicalin-1-like isoform X2 [Populus euphratica] |
| 8 |
Hb_009620_090 |
0.2505066835 |
- |
- |
hypothetical protein L484_000980 [Morus notabilis] |
| 9 |
Hb_001329_190 |
0.2510799307 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 10 |
Hb_000742_190 |
0.2519549663 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000544_030 |
0.2527628101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_074836_010 |
0.2531347462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000948_200 |
0.2545107877 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 14 |
Hb_025214_120 |
0.2590802159 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 15 |
Hb_161294_010 |
0.2627914109 |
- |
- |
PREDICTED: serine/threonine-protein kinase ppk15 isoform X2 [Jatropha curcas] |
| 16 |
Hb_010488_040 |
0.2632872601 |
- |
- |
PREDICTED: phosphomannomutase [Populus euphratica] |
| 17 |
Hb_001016_010 |
0.2694890899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002837_050 |
0.2703547714 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 19 |
Hb_001454_070 |
0.2709487449 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 20 |
Hb_010789_020 |
0.2733359115 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |